Probe CUST_39515_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39515_PI426222305 | JHI_St_60k_v1 | DMT400019061 | GGAAATCATAACTAATATTTCAACTGTTTGTGTTAAAACTAGTCCCTCCGATCAAACCTC |
All Microarray Probes Designed to Gene DMG402007388
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39515_PI426222305 | JHI_St_60k_v1 | DMT400019061 | GGAAATCATAACTAATATTTCAACTGTTTGTGTTAAAACTAGTCCCTCCGATCAAACCTC |
Microarray Signals from CUST_39515_PI426222305
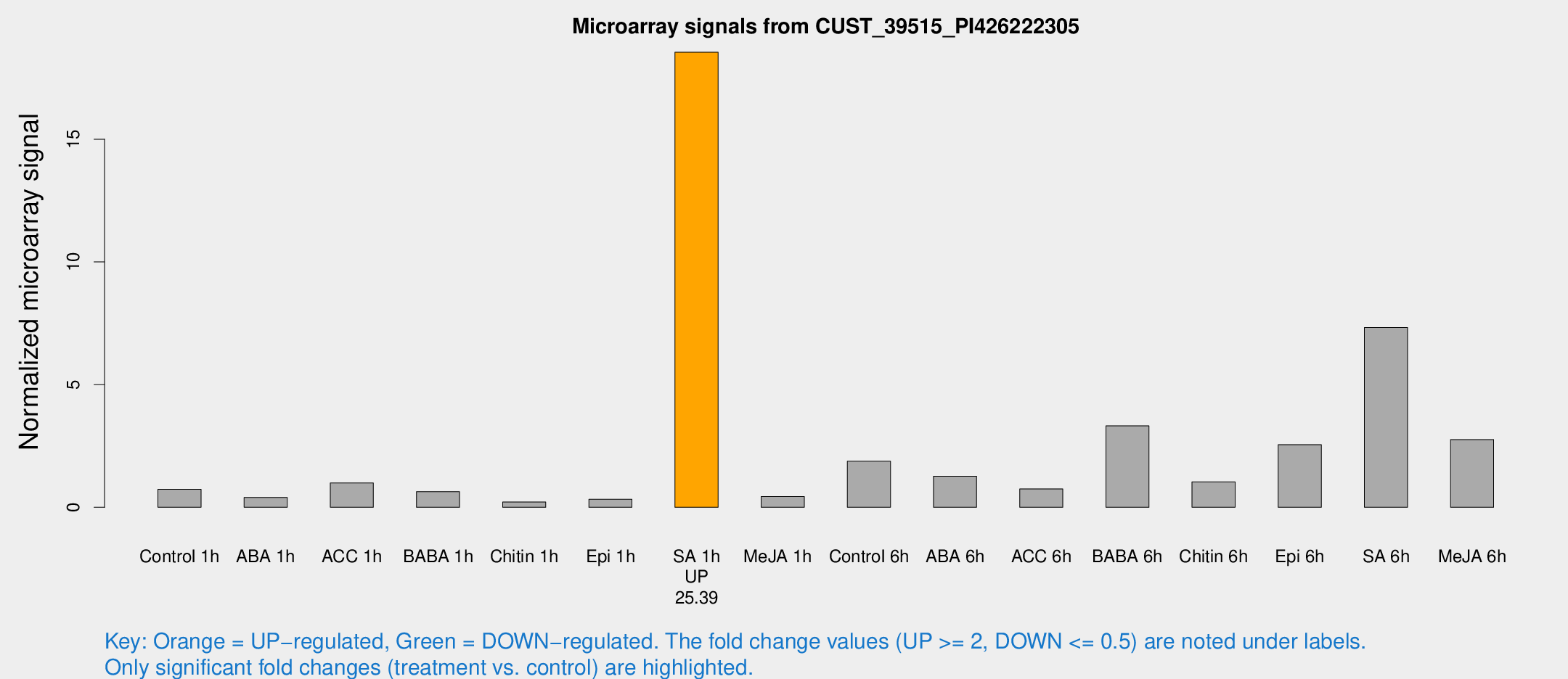
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 220.424 | 71.3358 | 0.730434 | 0.184356 |
| ABA 1h | 119.261 | 48.1589 | 0.403794 | 0.200398 |
| ACC 1h | 427.835 | 270.509 | 0.991297 | 0.798781 |
| BABA 1h | 200.806 | 69.5079 | 0.635531 | 0.24983 |
| Chitin 1h | 88.8835 | 48.1904 | 0.215618 | 0.310425 |
| Epi 1h | 99.4768 | 42.9315 | 0.325549 | 0.217953 |
| SA 1h | 5863.79 | 2028.03 | 18.5438 | 5.33873 |
| Me-JA 1h | 112.973 | 43.6929 | 0.437802 | 0.131768 |
| Control 6h | 667.147 | 270.102 | 1.87764 | 1.03834 |
| ABA 6h | 461.899 | 174.509 | 1.26858 | 0.709961 |
| ACC 6h | 238.129 | 21.5768 | 0.749709 | 0.167167 |
| BABA 6h | 1052.39 | 193.432 | 3.31739 | 0.4737 |
| Chitin 6h | 330.899 | 91.8208 | 1.03618 | 0.328801 |
| Epi 6h | 835.636 | 180.385 | 2.55422 | 0.491426 |
| SA 6h | 2107.24 | 454.075 | 7.32319 | 0.983981 |
| Me-JA 6h | 981.76 | 466.425 | 2.76227 | 1.71739 |
Source Transcript PGSC0003DMT400019061 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G80840.1 | +2 | 7e-56 | 189 | 121/281 (43%) | WRKY DNA-binding protein 40 | chr1:30383834-30385356 FORWARD LENGTH=302 |
