Probe CUST_39230_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39230_PI426222305 | JHI_St_60k_v1 | DMT400016722 | CTCAATTGTAGTGTTTCCTCATAACTCCCTCCATGTATTTTATATGTGTGTTGATGATCT |
All Microarray Probes Designed to Gene DMG400006538
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39230_PI426222305 | JHI_St_60k_v1 | DMT400016722 | CTCAATTGTAGTGTTTCCTCATAACTCCCTCCATGTATTTTATATGTGTGTTGATGATCT |
Microarray Signals from CUST_39230_PI426222305
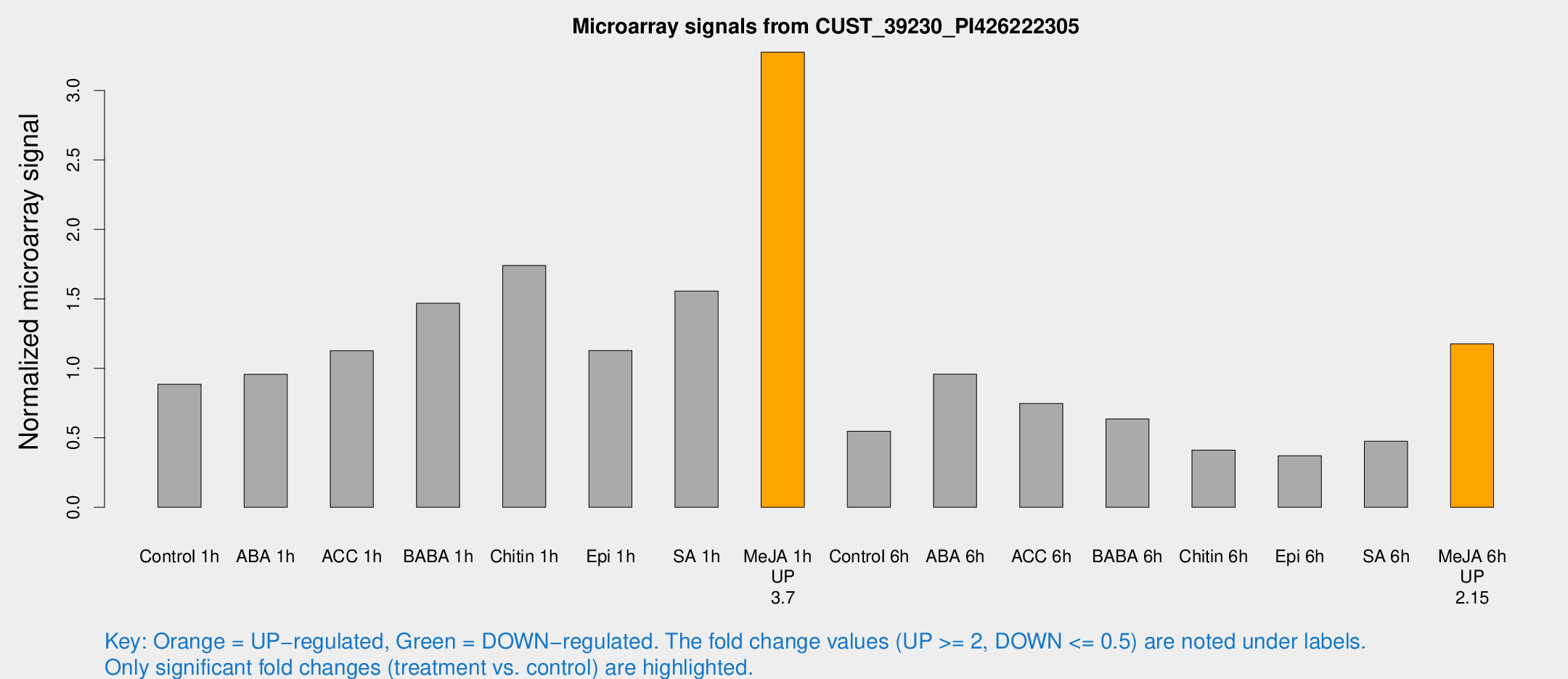
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 337.791 | 74.7923 | 0.885558 | 0.242344 |
| ABA 1h | 307.526 | 34.4493 | 0.957175 | 0.0578085 |
| ACC 1h | 421.46 | 49.8543 | 1.12626 | 0.200385 |
| BABA 1h | 532.981 | 109.075 | 1.46855 | 0.186538 |
| Chitin 1h | 566.544 | 52.195 | 1.74009 | 0.136256 |
| Epi 1h | 353.016 | 29.1244 | 1.1285 | 0.0919256 |
| SA 1h | 599.323 | 115.504 | 1.55652 | 0.273504 |
| Me-JA 1h | 991.693 | 166.16 | 3.27526 | 0.405905 |
| Control 6h | 211.839 | 51.5923 | 0.546923 | 0.106468 |
| ABA 6h | 386.79 | 86.7748 | 0.958435 | 0.18596 |
| ACC 6h | 310.75 | 29.8245 | 0.747578 | 0.0738928 |
| BABA 6h | 258.227 | 26.1782 | 0.635569 | 0.0789993 |
| Chitin 6h | 159.558 | 20.8731 | 0.410978 | 0.0535056 |
| Epi 6h | 151.972 | 16.4633 | 0.370974 | 0.0296636 |
| SA 6h | 174.542 | 31.2674 | 0.475643 | 0.0442315 |
| Me-JA 6h | 421.223 | 24.5537 | 1.17646 | 0.0989971 |
Source Transcript PGSC0003DMT400016722 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G36430.1 | +2 | 1e-47 | 174 | 124/419 (30%) | Plant protein of unknown function (DUF247) | chr2:15290211-15291557 FORWARD LENGTH=448 |
