Probe CUST_38291_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38291_PI426222305 | JHI_St_60k_v1 | DMT400067296 | ATGGACTACAGAACGACGCCGCAAGAGCTTTTCCTGTTTATTTGATATTTAATTATGTTT |
All Microarray Probes Designed to Gene DMG400026169
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38291_PI426222305 | JHI_St_60k_v1 | DMT400067296 | ATGGACTACAGAACGACGCCGCAAGAGCTTTTCCTGTTTATTTGATATTTAATTATGTTT |
Microarray Signals from CUST_38291_PI426222305
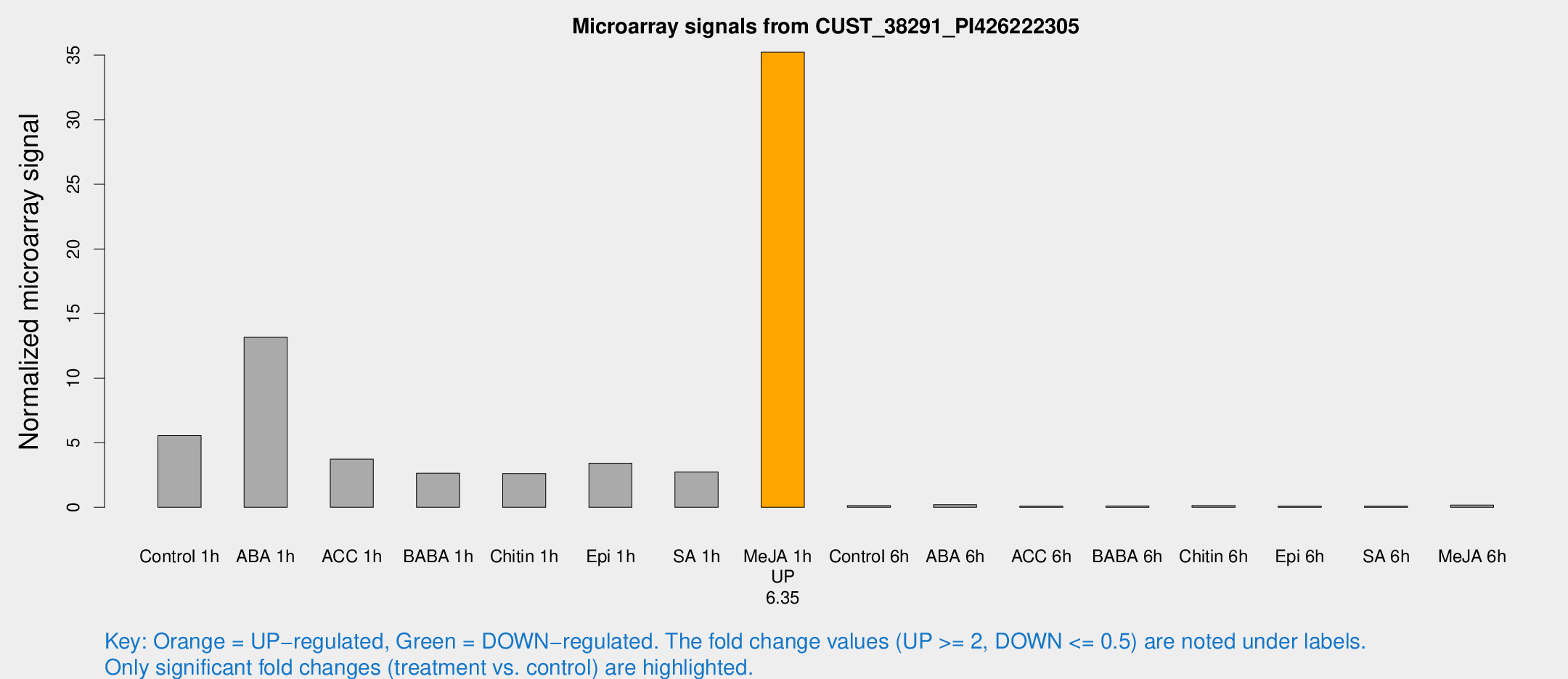
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3933.8 | 1158.15 | 5.5476 | 1.31657 |
| ABA 1h | 7571.46 | 437.521 | 13.1556 | 0.75956 |
| ACC 1h | 2564.79 | 429.775 | 3.73071 | 0.789634 |
| BABA 1h | 1752.36 | 393.789 | 2.64249 | 0.396878 |
| Chitin 1h | 1673.74 | 518.182 | 2.62113 | 0.793341 |
| Epi 1h | 1930.38 | 137.245 | 3.41294 | 0.339574 |
| SA 1h | 1914.46 | 406.81 | 2.73486 | 0.739267 |
| Me-JA 1h | 19045.4 | 2625.87 | 35.2259 | 4.66068 |
| Control 6h | 90.736 | 21.2357 | 0.130173 | 0.023464 |
| ABA 6h | 141.664 | 15.8438 | 0.202251 | 0.0136246 |
| ACC 6h | 66.5323 | 10.4487 | 0.0871876 | 0.0243984 |
| BABA 6h | 77.8384 | 12.11 | 0.104045 | 0.0200631 |
| Chitin 6h | 92.9172 | 10.1141 | 0.13278 | 0.0149326 |
| Epi 6h | 66.2765 | 8.61398 | 0.0889934 | 0.017744 |
| SA 6h | 53.5079 | 7.43824 | 0.0817189 | 0.0216501 |
| Me-JA 6h | 116.23 | 34.2486 | 0.166021 | 0.064012 |
Source Transcript PGSC0003DMT400067296 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G15248.1 | +2 | 3e-27 | 100 | 48/100 (48%) | B-box type zinc finger family protein | chr4:8708881-8709234 FORWARD LENGTH=117 |
