Probe CUST_38234_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38234_PI426222305 | JHI_St_60k_v1 | DMT400067321 | TGGCACACAGCACTGAAGAAGCGAACTAATTATAACTTGAGTGAAGGAAGTAAGAAATGC |
All Microarray Probes Designed to Gene DMG401026177
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38234_PI426222305 | JHI_St_60k_v1 | DMT400067321 | TGGCACACAGCACTGAAGAAGCGAACTAATTATAACTTGAGTGAAGGAAGTAAGAAATGC |
Microarray Signals from CUST_38234_PI426222305
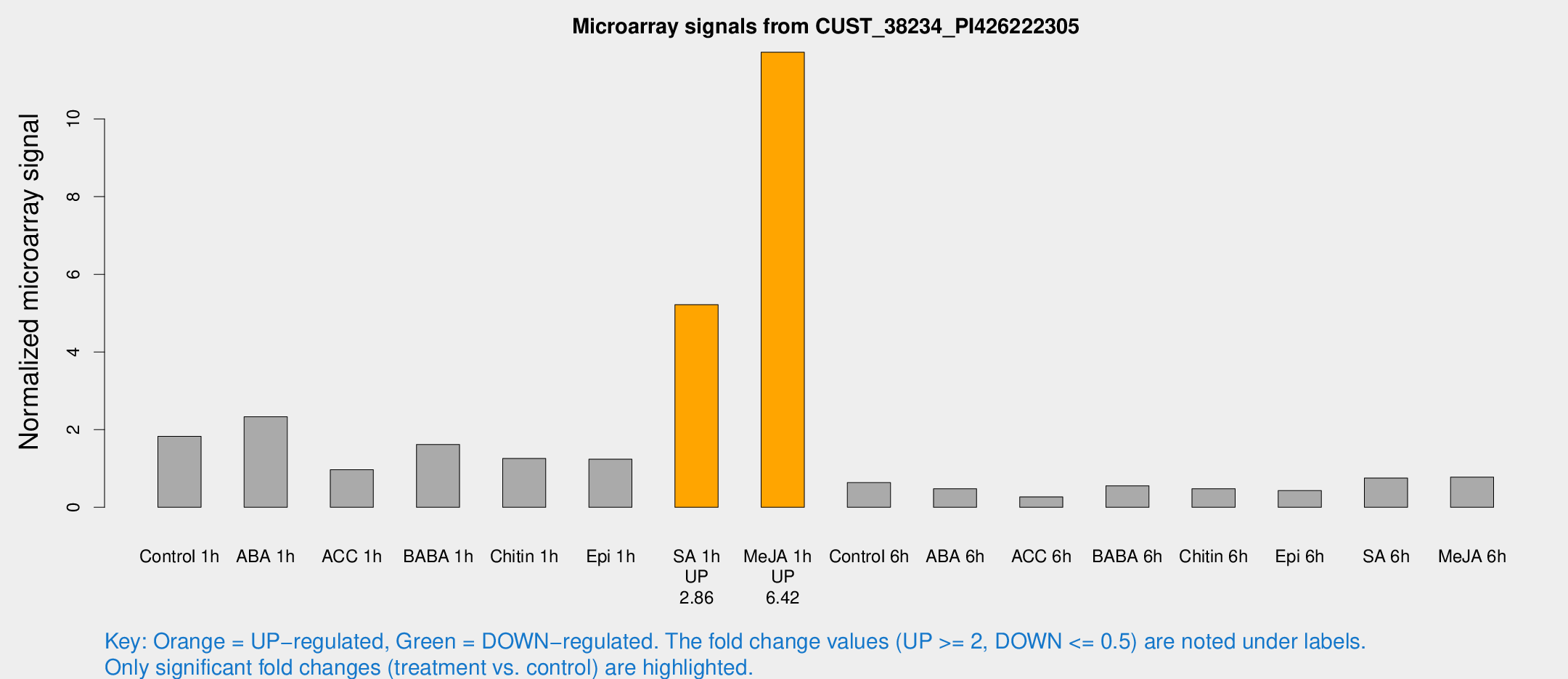
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 77.5227 | 15.4457 | 1.8257 | 0.234971 |
| ABA 1h | 88.8661 | 19.4904 | 2.33276 | 0.487965 |
| ACC 1h | 56.5163 | 27.1689 | 0.966601 | 0.697328 |
| BABA 1h | 63.9443 | 5.36121 | 1.61748 | 0.164163 |
| Chitin 1h | 46.9647 | 6.1204 | 1.25555 | 0.24886 |
| Epi 1h | 44.2469 | 4.1536 | 1.23775 | 0.148788 |
| SA 1h | 222.224 | 24.1002 | 5.21726 | 0.929812 |
| Me-JA 1h | 392.738 | 23.0264 | 11.7197 | 0.68507 |
| Control 6h | 32.2531 | 11.9983 | 0.634545 | 0.302064 |
| ABA 6h | 22.0152 | 5.26648 | 0.478717 | 0.103394 |
| ACC 6h | 13.0049 | 4.63938 | 0.26808 | 0.0980621 |
| BABA 6h | 27.9142 | 7.81304 | 0.554855 | 0.206945 |
| Chitin 6h | 25.7186 | 10.4773 | 0.480178 | 0.267276 |
| Epi 6h | 22.3612 | 6.70016 | 0.428873 | 0.21419 |
| SA 6h | 32.8949 | 8.13274 | 0.751416 | 0.136523 |
| Me-JA 6h | 33.1112 | 6.98839 | 0.778917 | 0.129049 |
Source Transcript PGSC0003DMT400067321 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G31180.1 | +1 | 1e-52 | 174 | 78/120 (65%) | myb domain protein 14 | chr2:13286806-13288175 REVERSE LENGTH=249 |
