Probe CUST_38222_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38222_PI426222305 | JHI_St_60k_v1 | DMT400067383 | GGAGACGTTCTGGTTCATCCTTTGTAACAAGTCATTTTCTATTCCAGATGGAAATATATT |
All Microarray Probes Designed to Gene DMG400026195
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38222_PI426222305 | JHI_St_60k_v1 | DMT400067383 | GGAGACGTTCTGGTTCATCCTTTGTAACAAGTCATTTTCTATTCCAGATGGAAATATATT |
Microarray Signals from CUST_38222_PI426222305
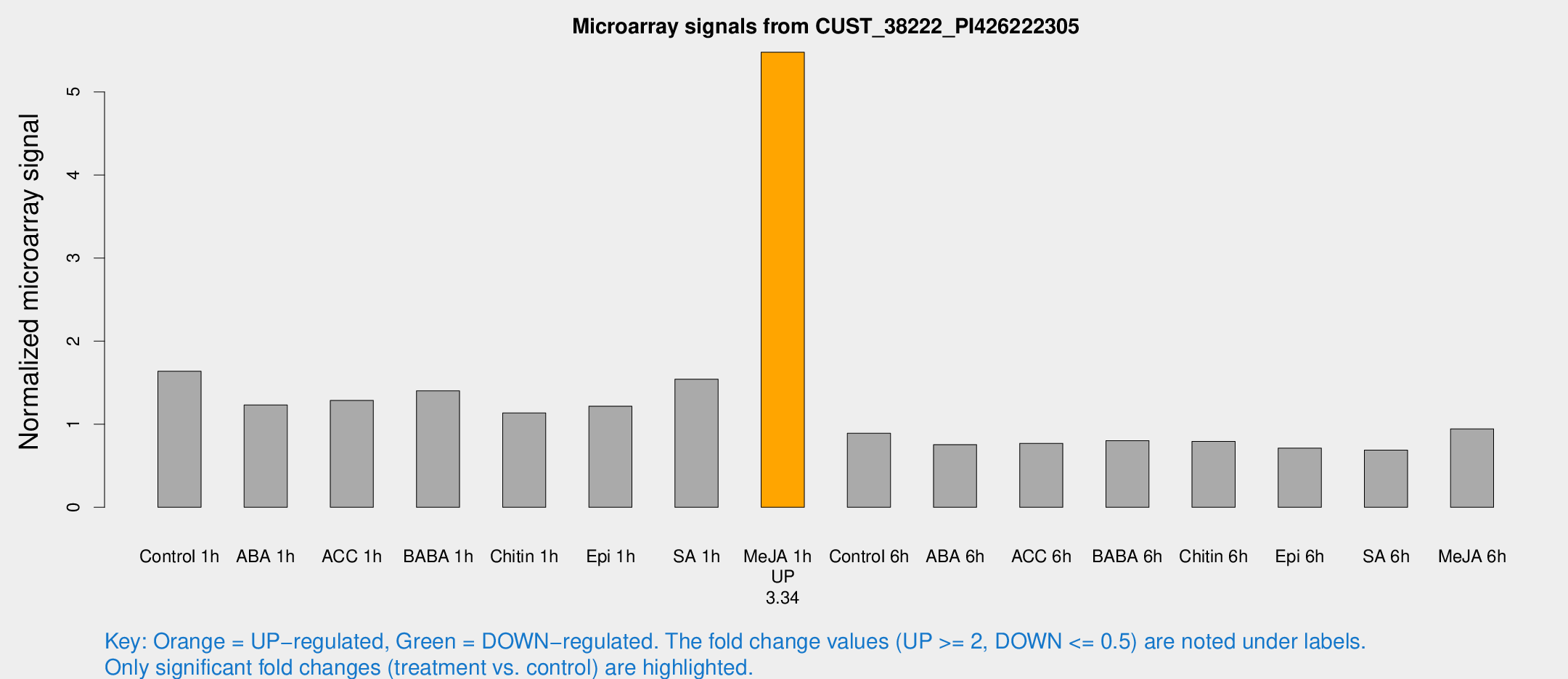
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8043.3 | 1685.31 | 1.63868 | 0.245389 |
| ABA 1h | 5273.09 | 805.588 | 1.23234 | 0.124645 |
| ACC 1h | 6716.39 | 1831.45 | 1.28658 | 0.29301 |
| BABA 1h | 6520.15 | 937.714 | 1.40174 | 0.0809323 |
| Chitin 1h | 4823.84 | 279.043 | 1.1358 | 0.0739873 |
| Epi 1h | 5119.7 | 827.966 | 1.21725 | 0.204816 |
| SA 1h | 7509.38 | 513.753 | 1.54312 | 0.0890945 |
| Me-JA 1h | 21349.1 | 2378.5 | 5.47846 | 0.3163 |
| Control 6h | 4443.19 | 925.459 | 0.891459 | 0.134491 |
| ABA 6h | 3823.03 | 406.448 | 0.754469 | 0.043564 |
| ACC 6h | 4247.25 | 596.014 | 0.77006 | 0.0444644 |
| BABA 6h | 4246.83 | 245.842 | 0.802619 | 0.0463439 |
| Chitin 6h | 3995.94 | 231.434 | 0.793812 | 0.0458358 |
| Epi 6h | 3813.1 | 339.432 | 0.711924 | 0.0708796 |
| SA 6h | 3437.88 | 813.253 | 0.688415 | 0.111645 |
| Me-JA 6h | 4493.83 | 528.163 | 0.944527 | 0.0545364 |
Source Transcript PGSC0003DMT400067383 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00390.1 | +2 | 1e-19 | 91 | 56/145 (39%) | DNA-binding storekeeper protein-related transcriptional regulator | chr4:171650-172744 REVERSE LENGTH=364 |
