Probe CUST_38031_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38031_PI426222305 | JHI_St_60k_v1 | DMT400081432 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
All Microarray Probes Designed to Gene DMG400031834
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38056_PI426222305 | JHI_St_60k_v1 | DMT400081428 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_38031_PI426222305 | JHI_St_60k_v1 | DMT400081432 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_38054_PI426222305 | JHI_St_60k_v1 | DMT400081430 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_37974_PI426222305 | JHI_St_60k_v1 | DMT400081431 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_37975_PI426222305 | JHI_St_60k_v1 | DMT400081429 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
Microarray Signals from CUST_38031_PI426222305
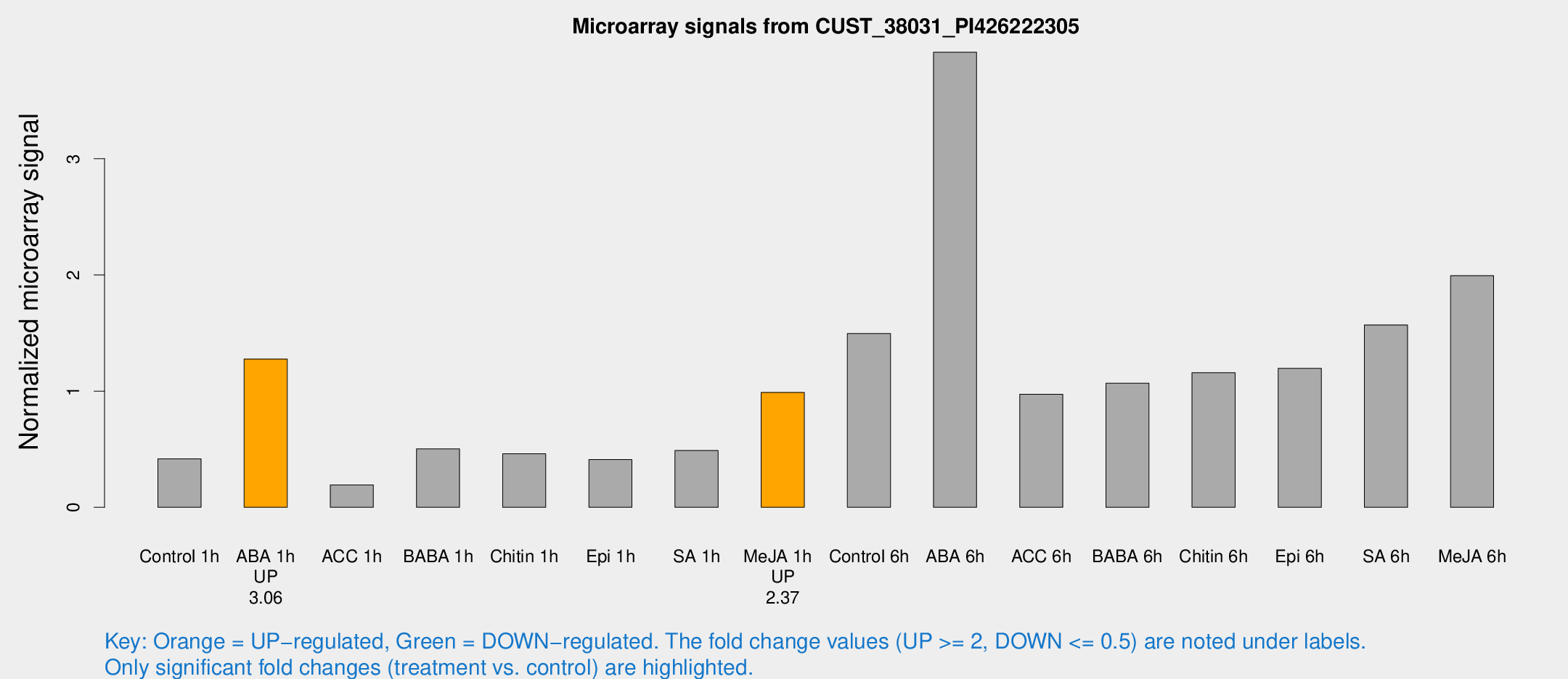
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 149.521 | 25.5615 | 0.416522 | 0.0895387 |
| ABA 1h | 411.939 | 96.8564 | 1.27613 | 0.208363 |
| ACC 1h | 86.5032 | 33.3286 | 0.192148 | 0.105047 |
| BABA 1h | 174.704 | 32.0753 | 0.502438 | 0.0508932 |
| Chitin 1h | 144.016 | 9.06516 | 0.460487 | 0.0288944 |
| Epi 1h | 127.846 | 23.6964 | 0.410777 | 0.0697576 |
| SA 1h | 181.196 | 35.3616 | 0.488388 | 0.0865741 |
| Me-JA 1h | 283.302 | 28.5094 | 0.989095 | 0.065846 |
| Control 6h | 535.947 | 96.4603 | 1.49561 | 0.203231 |
| ABA 6h | 1499.67 | 294.466 | 3.91682 | 0.62818 |
| ACC 6h | 389.621 | 29.0602 | 0.973447 | 0.17004 |
| BABA 6h | 418.412 | 39.5235 | 1.06795 | 0.141815 |
| Chitin 6h | 432.407 | 43.4823 | 1.15829 | 0.148346 |
| Epi 6h | 472.601 | 47.6743 | 1.19541 | 0.0971992 |
| SA 6h | 541.986 | 41.4736 | 1.56933 | 0.0913741 |
| Me-JA 6h | 709.196 | 125.664 | 1.99344 | 0.206597 |
Source Transcript PGSC0003DMT400081432 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G32150.1 | +2 | 5e-84 | 266 | 131/196 (67%) | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein | chr2:13659095-13660531 FORWARD LENGTH=263 |
