Probe CUST_38012_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38012_PI426222305 | JHI_St_60k_v1 | DMT400081405 | AATAATGCATTGTATAGTTTGAGAATAGTGCTCAAGGAAAGGCCGTTGCACAGAGCATTT |
All Microarray Probes Designed to Gene DMG400031821
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38042_PI426222305 | JHI_St_60k_v1 | DMT400081406 | GATGAAGAAGGATCCTTGCATTAGGGGAGAAGCTAAACATAGATTTCAACAAGTTCAAGA |
| CUST_38012_PI426222305 | JHI_St_60k_v1 | DMT400081405 | AATAATGCATTGTATAGTTTGAGAATAGTGCTCAAGGAAAGGCCGTTGCACAGAGCATTT |
Microarray Signals from CUST_38012_PI426222305
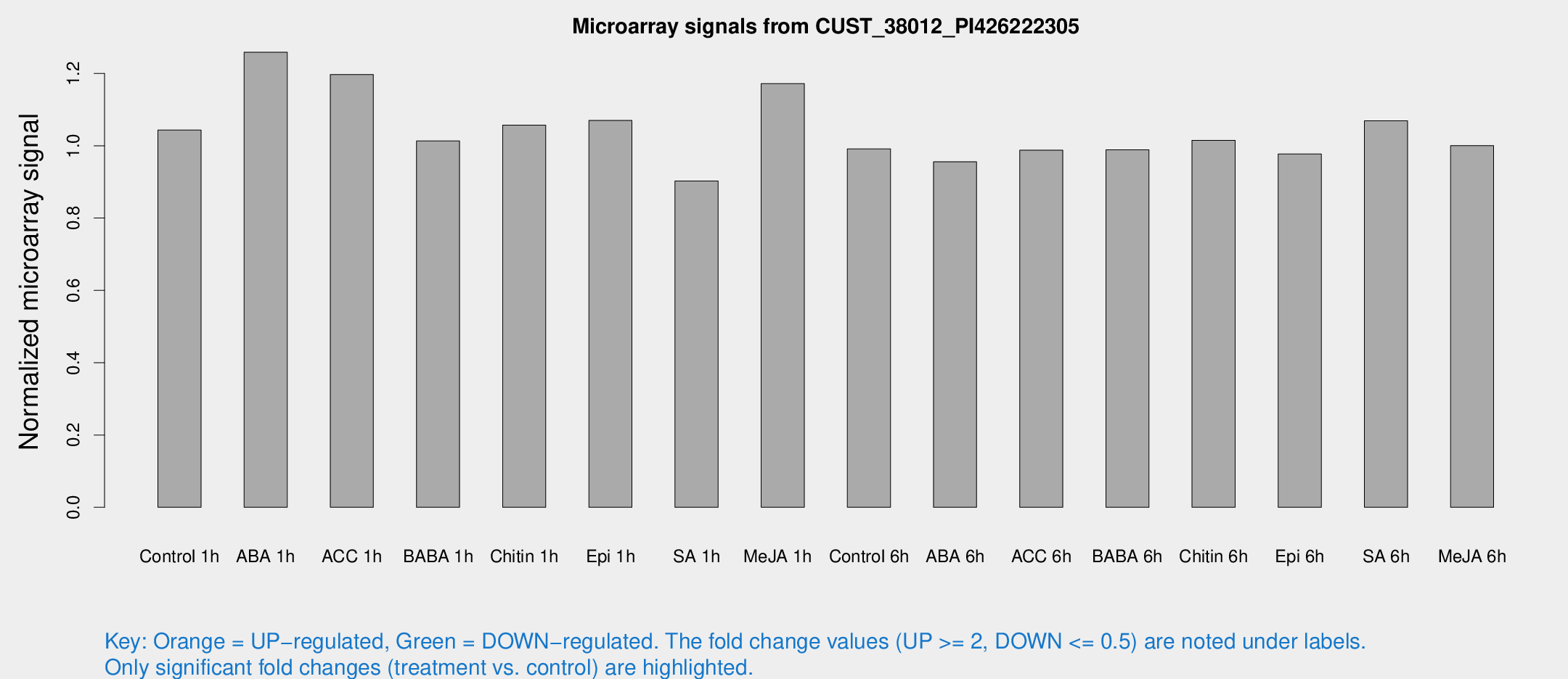
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.12859 | 3.09786 | 1.04362 | 0.541625 |
| ABA 1h | 6.51135 | 3.05634 | 1.25868 | 0.601263 |
| ACC 1h | 7.51486 | 3.41816 | 1.19713 | 0.59593 |
| BABA 1h | 5.68903 | 3.30624 | 1.01319 | 0.587641 |
| Chitin 1h | 5.47147 | 3.18644 | 1.05725 | 0.602869 |
| Epi 1h | 5.34377 | 3.1023 | 1.06999 | 0.613812 |
| SA 1h | 5.39321 | 3.12552 | 0.902497 | 0.522622 |
| Me-JA 1h | 5.58223 | 3.24577 | 1.17172 | 0.678532 |
| Control 6h | 5.77906 | 3.35448 | 0.991324 | 0.575032 |
| ABA 6h | 5.90887 | 3.42569 | 0.955515 | 0.553609 |
| ACC 6h | 6.7291 | 3.98815 | 0.987809 | 0.572693 |
| BABA 6h | 6.45273 | 3.74985 | 0.988586 | 0.572676 |
| Chitin 6h | 6.28335 | 3.63929 | 1.01515 | 0.587796 |
| Epi 6h | 6.46867 | 3.78408 | 0.977223 | 0.56587 |
| SA 6h | 6.15028 | 3.49739 | 1.06883 | 0.608133 |
| Me-JA 6h | 5.79633 | 3.35951 | 1.00042 | 0.579305 |
Source Transcript PGSC0003DMT400081405 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G56300.1 | +1 | 6e-12 | 61 | 31/63 (49%) | Chaperone DnaJ-domain superfamily protein | chr1:21079022-21080168 REVERSE LENGTH=156 |
