Probe CUST_37975_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37975_PI426222305 | JHI_St_60k_v1 | DMT400081429 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
All Microarray Probes Designed to Gene DMG400031834
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38056_PI426222305 | JHI_St_60k_v1 | DMT400081428 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_38054_PI426222305 | JHI_St_60k_v1 | DMT400081430 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_37975_PI426222305 | JHI_St_60k_v1 | DMT400081429 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_38031_PI426222305 | JHI_St_60k_v1 | DMT400081432 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
| CUST_37974_PI426222305 | JHI_St_60k_v1 | DMT400081431 | AAGCCACGTGAAGGTGGTTTTTTCGAGAACTTTTGTGTTGATTCGAGATAAGCTCAACAA |
Microarray Signals from CUST_37975_PI426222305
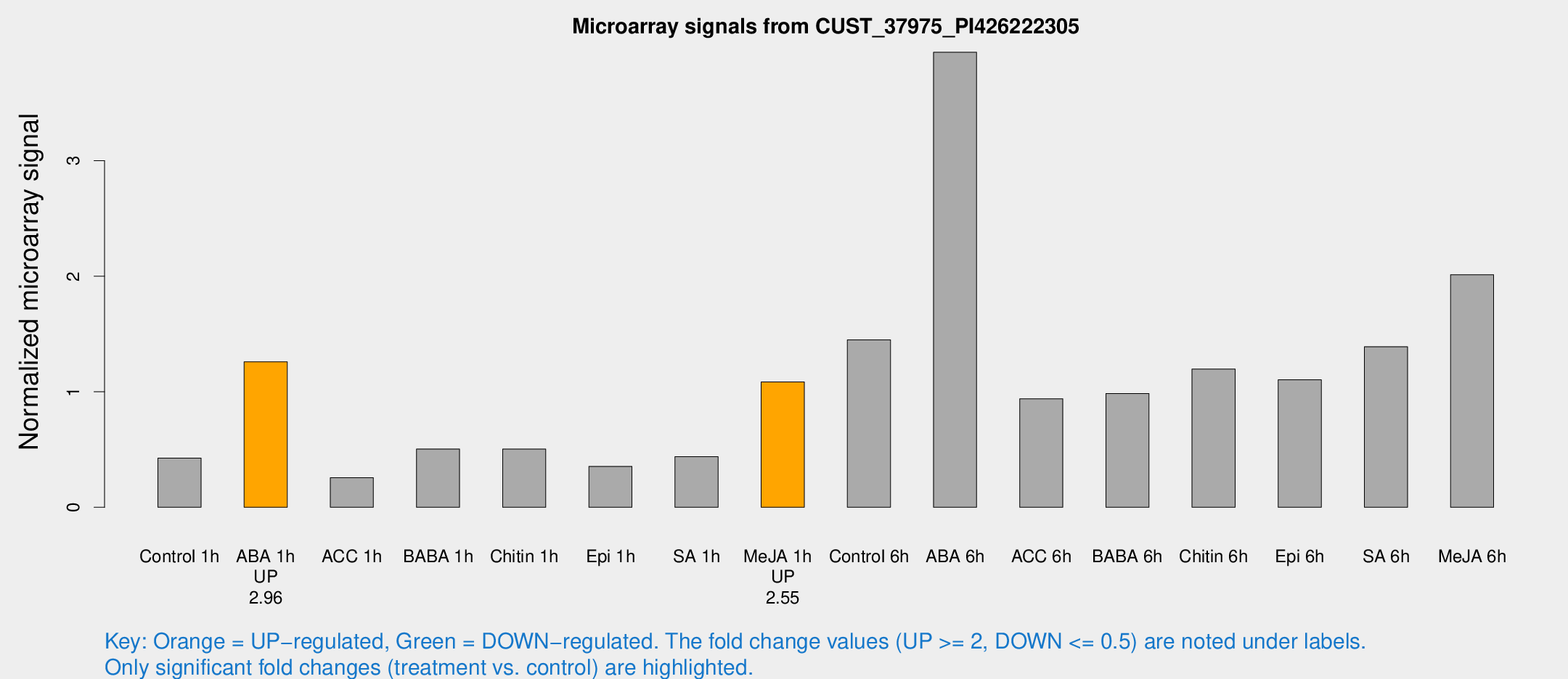
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 150.334 | 23.4056 | 0.425882 | 0.0793929 |
| ABA 1h | 398.71 | 84.6666 | 1.25884 | 0.171253 |
| ACC 1h | 92.8139 | 15.4184 | 0.256134 | 0.0301306 |
| BABA 1h | 170.974 | 24.9634 | 0.504618 | 0.0315448 |
| Chitin 1h | 155.719 | 9.73036 | 0.503937 | 0.0314422 |
| Epi 1h | 115.922 | 32.9664 | 0.354257 | 0.114993 |
| SA 1h | 159.19 | 27.4158 | 0.439077 | 0.0725437 |
| Me-JA 1h | 304.654 | 18.0097 | 1.08536 | 0.0745408 |
| Control 6h | 520.255 | 111.446 | 1.44989 | 0.222465 |
| ABA 6h | 1475 | 244.635 | 3.93933 | 0.457498 |
| ACC 6h | 372.868 | 33.4537 | 0.939561 | 0.183578 |
| BABA 6h | 382.349 | 37.8589 | 0.985356 | 0.138521 |
| Chitin 6h | 444.748 | 58.4418 | 1.19742 | 0.163879 |
| Epi 6h | 428.949 | 28.2257 | 1.10352 | 0.0909837 |
| SA 6h | 475.754 | 40.9369 | 1.39001 | 0.0811758 |
| Me-JA 6h | 724.134 | 167.881 | 2.01385 | 0.294889 |
Source Transcript PGSC0003DMT400081429 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G32150.1 | +2 | 1e-76 | 248 | 131/233 (56%) | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein | chr2:13659095-13660531 FORWARD LENGTH=263 |
