Probe CUST_37843_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37843_PI426222305 | JHI_St_60k_v1 | DMT400005709 | CTAACTGTACAGTGTTAGGAAGGGAAGAATTAAAAGTGTCATAAATACATGCCTTGTCAA |
All Microarray Probes Designed to Gene DMG400002235
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37843_PI426222305 | JHI_St_60k_v1 | DMT400005709 | CTAACTGTACAGTGTTAGGAAGGGAAGAATTAAAAGTGTCATAAATACATGCCTTGTCAA |
Microarray Signals from CUST_37843_PI426222305
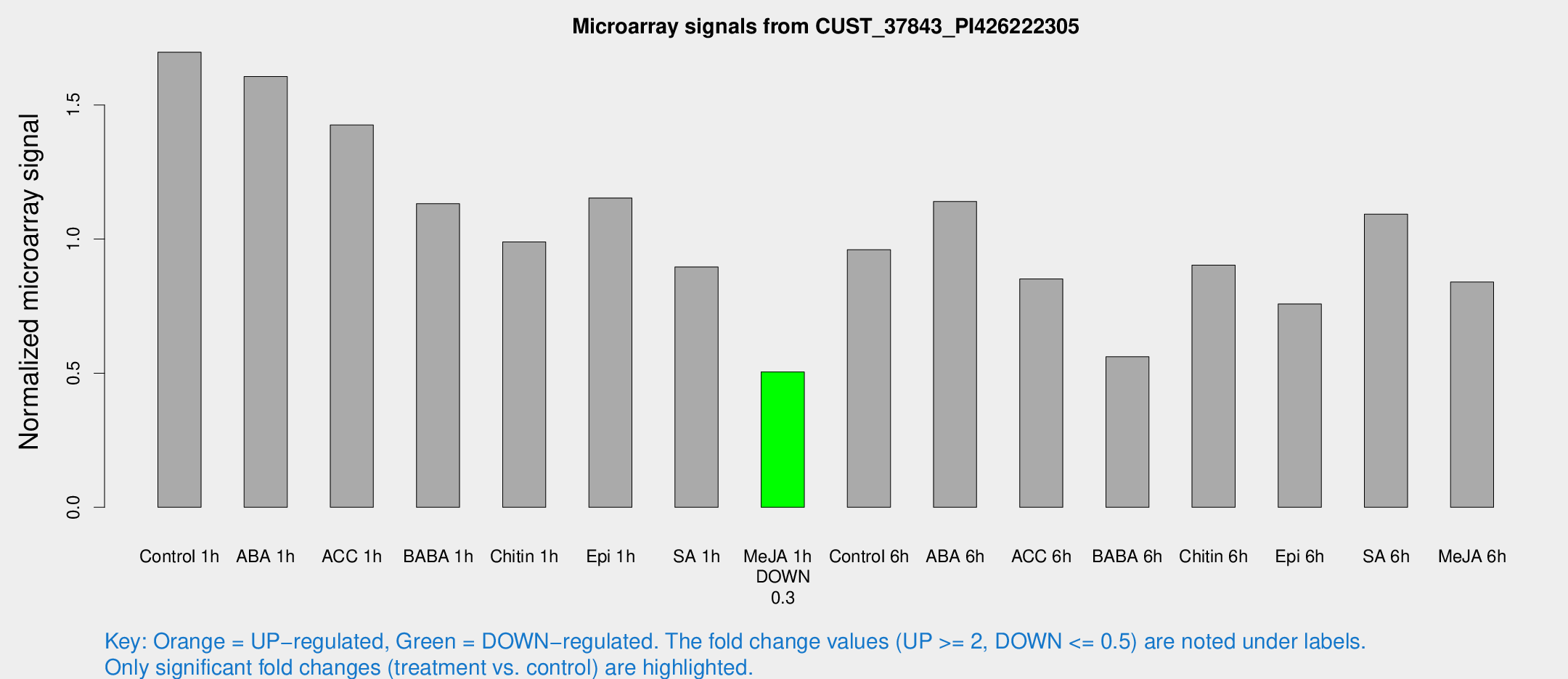
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 224.946 | 17.5127 | 1.69671 | 0.100389 |
| ABA 1h | 192.745 | 34.1328 | 1.60598 | 0.298629 |
| ACC 1h | 197.639 | 30.0849 | 1.42525 | 0.140886 |
| BABA 1h | 144.23 | 8.89058 | 1.1321 | 0.179107 |
| Chitin 1h | 119.934 | 18.5991 | 0.989546 | 0.172699 |
| Epi 1h | 132.008 | 8.17216 | 1.15327 | 0.0713682 |
| SA 1h | 121.409 | 7.59162 | 0.896113 | 0.0560209 |
| Me-JA 1h | 55.1366 | 6.81637 | 0.504922 | 0.0433307 |
| Control 6h | 131.276 | 25.0786 | 0.960576 | 0.15035 |
| ABA 6h | 160.081 | 9.78073 | 1.14002 | 0.069544 |
| ACC 6h | 130.428 | 14.9252 | 0.851255 | 0.143935 |
| BABA 6h | 86.9709 | 19.9663 | 0.5611 | 0.117585 |
| Chitin 6h | 127.266 | 8.213 | 0.903056 | 0.067699 |
| Epi 6h | 112.927 | 7.40257 | 0.758202 | 0.0495587 |
| SA 6h | 145.457 | 20.3311 | 1.09294 | 0.0682207 |
| Me-JA 6h | 112.061 | 13.6121 | 0.839887 | 0.0539606 |
Source Transcript PGSC0003DMT400005709 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G25810.1 | +1 | 9e-52 | 173 | 104/167 (62%) | Integrase-type DNA-binding superfamily protein | chr5:8986976-8987632 REVERSE LENGTH=218 |
