Probe CUST_37588_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37588_PI426222305 | JHI_St_60k_v1 | DMT400049493 | CAGCAGATGAAATAGCTTTTCTTGCATGTCATGTAGTTTCACTGTTCCATAGATAATTTC |
All Microarray Probes Designed to Gene DMG400019232
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37588_PI426222305 | JHI_St_60k_v1 | DMT400049493 | CAGCAGATGAAATAGCTTTTCTTGCATGTCATGTAGTTTCACTGTTCCATAGATAATTTC |
Microarray Signals from CUST_37588_PI426222305
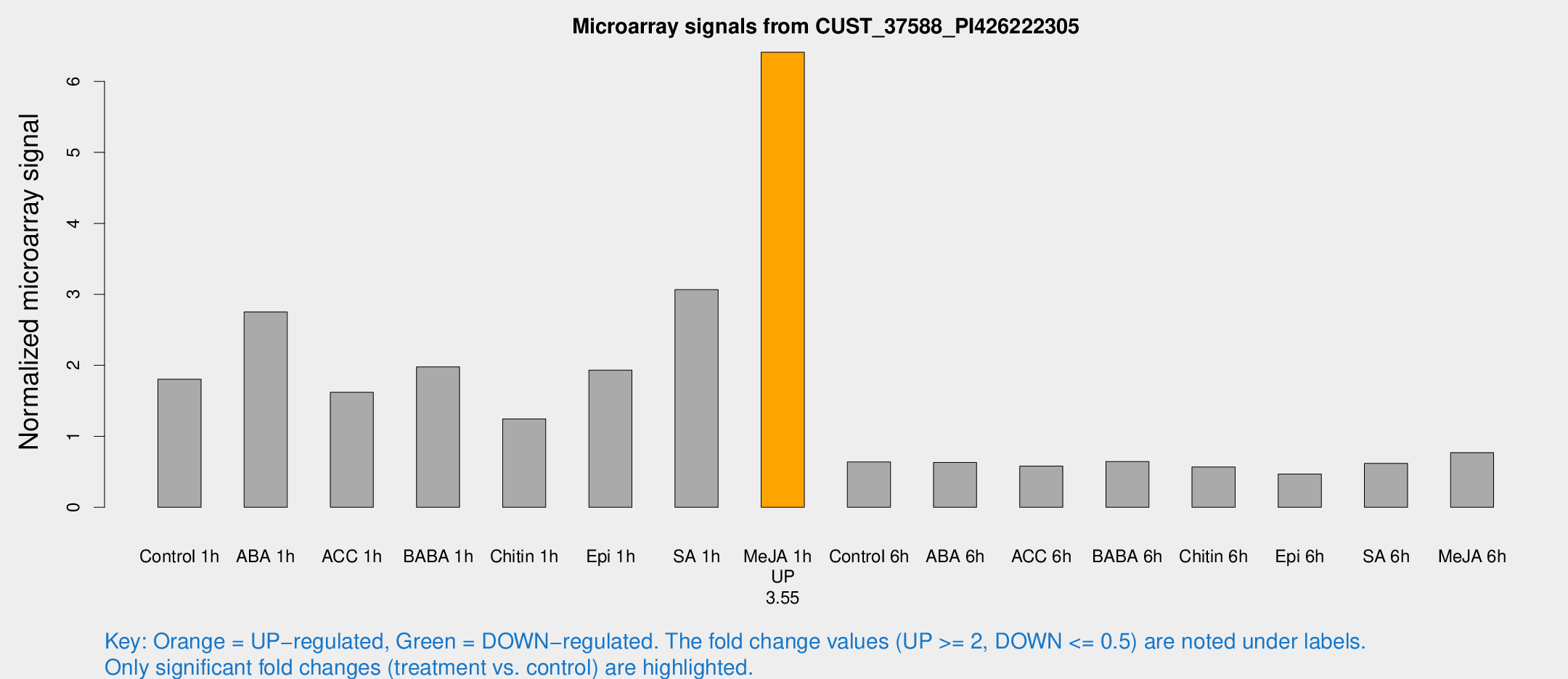
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2943.72 | 487.058 | 1.80369 | 0.192295 |
| ABA 1h | 4018 | 724.786 | 2.75195 | 0.368027 |
| ACC 1h | 2854.14 | 737.219 | 1.61972 | 0.361731 |
| BABA 1h | 3153.85 | 566.24 | 1.97947 | 0.192789 |
| Chitin 1h | 1814.5 | 251.845 | 1.24554 | 0.0911035 |
| Epi 1h | 2670.41 | 154.652 | 1.93344 | 0.111648 |
| SA 1h | 5152.29 | 795.902 | 3.06671 | 0.37428 |
| Me-JA 1h | 8485.47 | 1197.81 | 6.41073 | 0.391679 |
| Control 6h | 1087.06 | 248.884 | 0.638906 | 0.111252 |
| ABA 6h | 1080.64 | 111.522 | 0.631911 | 0.0365329 |
| ACC 6h | 1066.93 | 87.6708 | 0.580665 | 0.0339361 |
| BABA 6h | 1152.4 | 66.8182 | 0.645035 | 0.0375678 |
| Chitin 6h | 965.232 | 56.0065 | 0.568083 | 0.0328598 |
| Epi 6h | 857.394 | 113.66 | 0.468748 | 0.0426542 |
| SA 6h | 1007.29 | 182.519 | 0.618693 | 0.0624424 |
| Me-JA 6h | 1233.71 | 126.817 | 0.770308 | 0.0822274 |
Source Transcript PGSC0003DMT400049493 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G48150.1 | +2 | 2e-169 | 498 | 242/372 (65%) | GRAS family transcription factor | chr5:19522497-19524053 REVERSE LENGTH=490 |
