Probe CUST_37524_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37524_PI426222305 | JHI_St_60k_v1 | DMT400078930 | GGTGCATTTTCCTTGGAGTTTAGTGTGGAAATAGAGTAGTATGATTATGTTTTTTGACTC |
All Microarray Probes Designed to Gene DMG400030713
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37524_PI426222305 | JHI_St_60k_v1 | DMT400078930 | GGTGCATTTTCCTTGGAGTTTAGTGTGGAAATAGAGTAGTATGATTATGTTTTTTGACTC |
| CUST_37451_PI426222305 | JHI_St_60k_v1 | DMT400078929 | GGTGCATTTTCCTTGGAGTTTAGTGTGGAAATAGAGTAGTATGATTATGTTTTTTGACTC |
Microarray Signals from CUST_37524_PI426222305
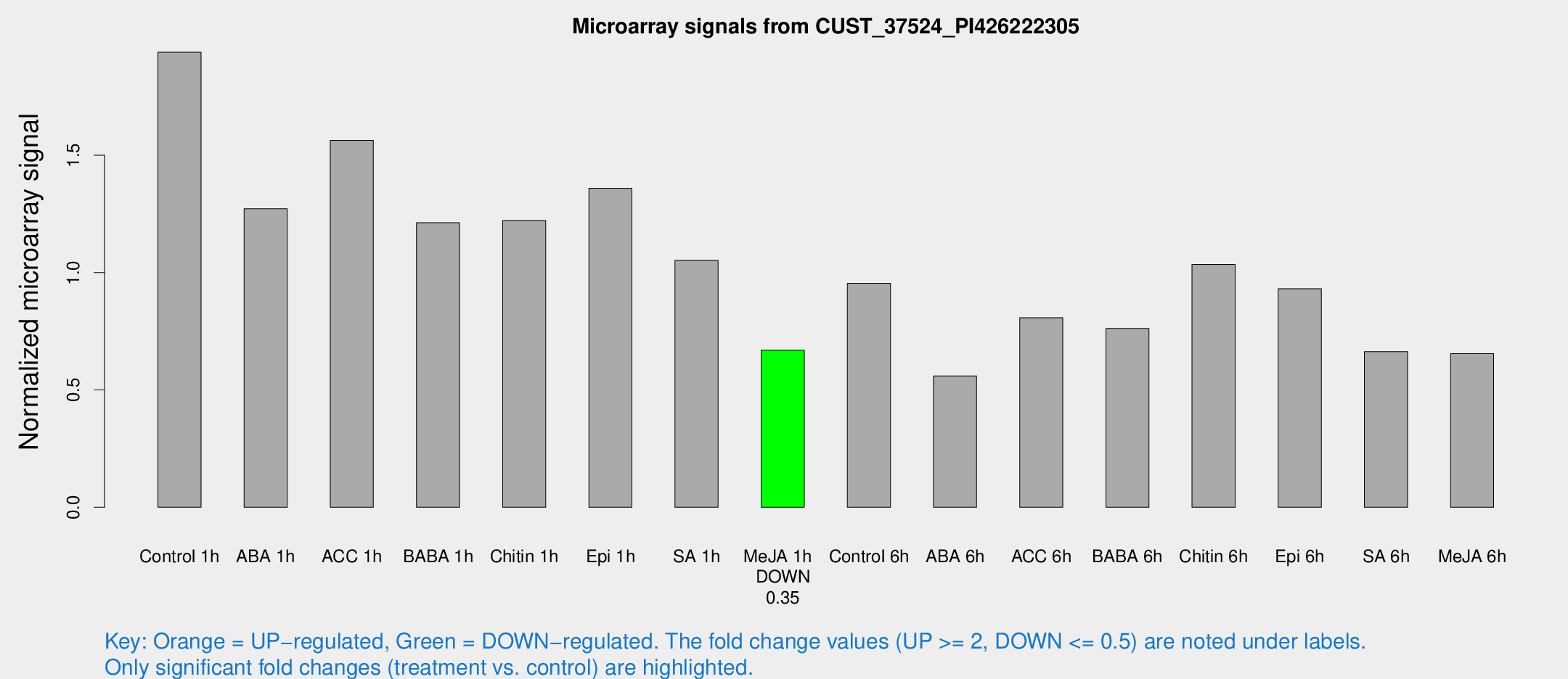
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1479.3 | 248.871 | 1.93933 | 0.212225 |
| ABA 1h | 842.126 | 88.0822 | 1.27224 | 0.0857985 |
| ACC 1h | 1269.75 | 312.893 | 1.56392 | 0.299394 |
| BABA 1h | 902.789 | 172.773 | 1.21251 | 0.134704 |
| Chitin 1h | 827.566 | 111.019 | 1.22196 | 0.170808 |
| Epi 1h | 873.994 | 50.7513 | 1.35932 | 0.078683 |
| SA 1h | 803.021 | 48.4629 | 1.05196 | 0.0609152 |
| Me-JA 1h | 404.877 | 23.6581 | 0.669699 | 0.0550558 |
| Control 6h | 776.6 | 214.475 | 0.954985 | 0.231705 |
| ABA 6h | 448.773 | 58.2667 | 0.55994 | 0.0714355 |
| ACC 6h | 701.252 | 105.97 | 0.807551 | 0.046915 |
| BABA 6h | 647.189 | 97.9804 | 0.762181 | 0.099875 |
| Chitin 6h | 817.179 | 47.4387 | 1.03502 | 0.0599858 |
| Epi 6h | 790.72 | 98.7864 | 0.931265 | 0.181869 |
| SA 6h | 494.047 | 60.1755 | 0.66356 | 0.0387256 |
| Me-JA 6h | 518.114 | 128.269 | 0.654782 | 0.127592 |
Source Transcript PGSC0003DMT400078930 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G20410.1 | +2 | 0.0 | 699 | 334/417 (80%) | monogalactosyldiacylglycerol synthase 2 | chr5:6896765-6898581 FORWARD LENGTH=468 |
