Probe CUST_37522_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37522_PI426222305 | JHI_St_60k_v1 | DMT400078971 | CCTATAAAGTGAGGATTGAGGAGGGGAGAGTGTATACAAACCTTATCCATACTTTGTTGA |
All Microarray Probes Designed to Gene DMG400030731
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37522_PI426222305 | JHI_St_60k_v1 | DMT400078971 | CCTATAAAGTGAGGATTGAGGAGGGGAGAGTGTATACAAACCTTATCCATACTTTGTTGA |
| CUST_37443_PI426222305 | JHI_St_60k_v1 | DMT400078972 | GCACAAAAACTTACTATCCTTTTCACCATTCTCCTTATGGTTATTGCTGCTCATGACAAT |
Microarray Signals from CUST_37522_PI426222305
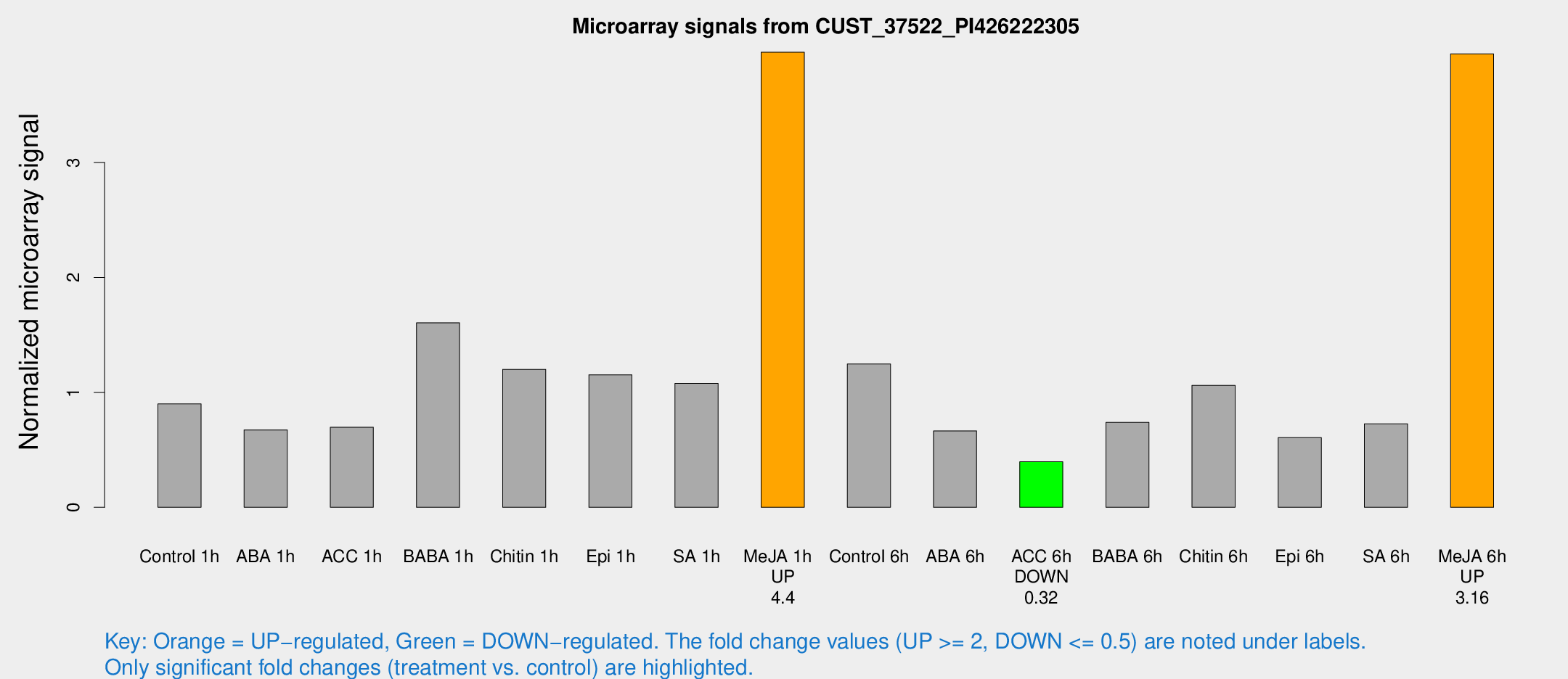
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 56.6753 | 11.7816 | 0.899451 | 0.241323 |
| ABA 1h | 38.4764 | 11.1774 | 0.673526 | 0.144744 |
| ACC 1h | 45.428 | 11.4573 | 0.696695 | 0.173067 |
| BABA 1h | 96.3107 | 18.8099 | 1.60567 | 0.18925 |
| Chitin 1h | 64.6476 | 5.1446 | 1.20057 | 0.126461 |
| Epi 1h | 63.0562 | 13.2862 | 1.15319 | 0.276539 |
| SA 1h | 68.3203 | 12.3288 | 1.07835 | 0.19019 |
| Me-JA 1h | 195.924 | 21.3559 | 3.95988 | 0.487054 |
| Control 6h | 85.1861 | 25.8575 | 1.24696 | 0.397859 |
| ABA 6h | 44.0616 | 8.56646 | 0.664725 | 0.171903 |
| ACC 6h | 28.6164 | 5.93863 | 0.396071 | 0.103717 |
| BABA 6h | 56.1572 | 21.0205 | 0.738563 | 0.269549 |
| Chitin 6h | 68.4567 | 7.69199 | 1.06052 | 0.149494 |
| Epi 6h | 46.621 | 14.5032 | 0.606759 | 0.311833 |
| SA 6h | 47.3974 | 13.5079 | 0.725988 | 0.352801 |
| Me-JA 6h | 245.084 | 45.7416 | 3.94518 | 0.538881 |
Source Transcript PGSC0003DMT400078971 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
