Probe CUST_37451_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37451_PI426222305 | JHI_St_60k_v1 | DMT400078929 | GGTGCATTTTCCTTGGAGTTTAGTGTGGAAATAGAGTAGTATGATTATGTTTTTTGACTC |
All Microarray Probes Designed to Gene DMG400030713
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37451_PI426222305 | JHI_St_60k_v1 | DMT400078929 | GGTGCATTTTCCTTGGAGTTTAGTGTGGAAATAGAGTAGTATGATTATGTTTTTTGACTC |
| CUST_37524_PI426222305 | JHI_St_60k_v1 | DMT400078930 | GGTGCATTTTCCTTGGAGTTTAGTGTGGAAATAGAGTAGTATGATTATGTTTTTTGACTC |
Microarray Signals from CUST_37451_PI426222305
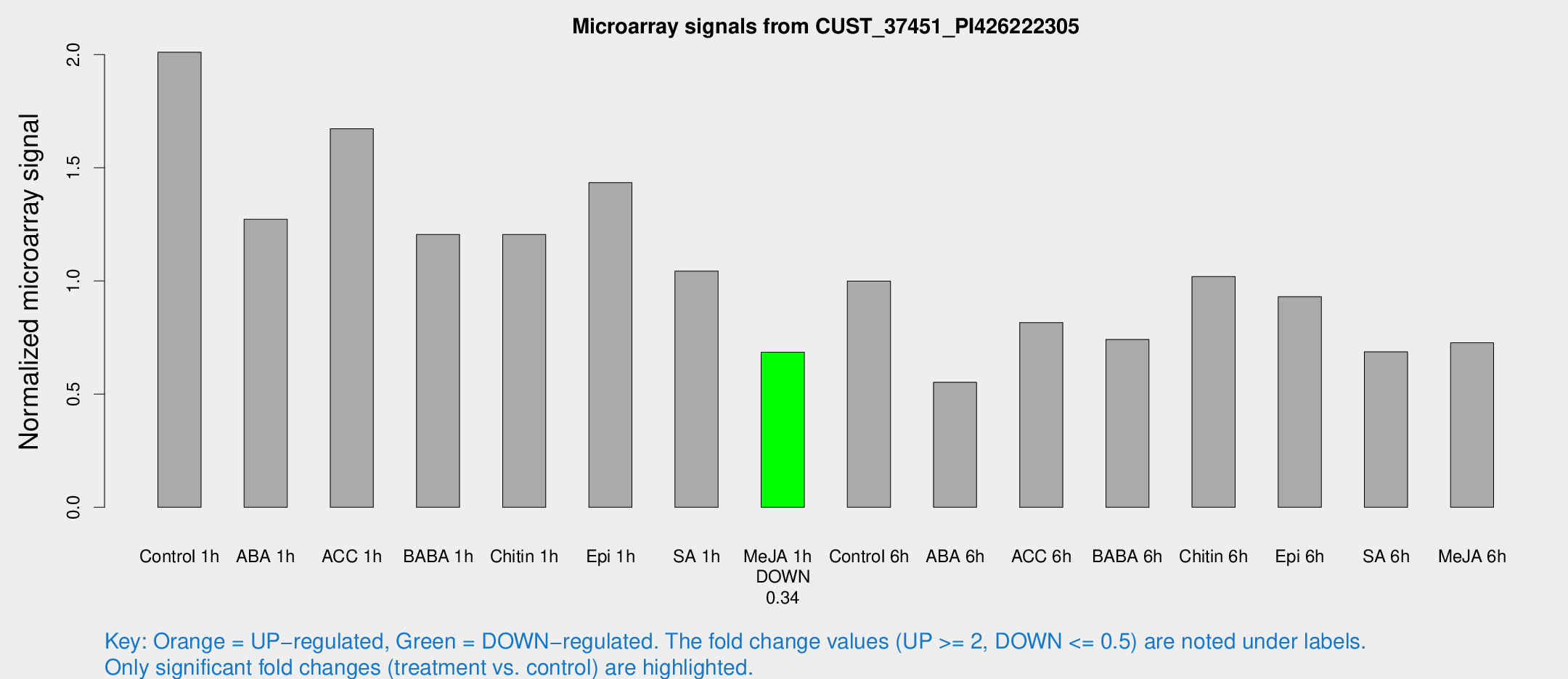
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1633.14 | 247.162 | 2.01063 | 0.194668 |
| ABA 1h | 893.998 | 51.8144 | 1.27231 | 0.0735625 |
| ACC 1h | 1435.18 | 330.14 | 1.67268 | 0.27716 |
| BABA 1h | 978.783 | 221.958 | 1.20558 | 0.190004 |
| Chitin 1h | 875.419 | 122.942 | 1.20543 | 0.164253 |
| Epi 1h | 989.992 | 79.638 | 1.43415 | 0.0874548 |
| SA 1h | 864.518 | 111.231 | 1.04362 | 0.0757995 |
| Me-JA 1h | 444.473 | 25.875 | 0.685741 | 0.0398485 |
| Control 6h | 871.006 | 241.693 | 0.999426 | 0.245216 |
| ABA 6h | 470.86 | 48.5622 | 0.552572 | 0.0599964 |
| ACC 6h | 767.627 | 140.659 | 0.816315 | 0.059034 |
| BABA 6h | 692.109 | 148.032 | 0.741628 | 0.153486 |
| Chitin 6h | 863.639 | 55.7114 | 1.01969 | 0.0956396 |
| Epi 6h | 838.007 | 77.9575 | 0.930156 | 0.11357 |
| SA 6h | 552.572 | 84.072 | 0.686916 | 0.0398905 |
| Me-JA 6h | 610.343 | 148.201 | 0.726705 | 0.124536 |
Source Transcript PGSC0003DMT400078929 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G20410.1 | +3 | 0.0 | 612 | 292/367 (80%) | monogalactosyldiacylglycerol synthase 2 | chr5:6896765-6898581 FORWARD LENGTH=468 |
