Probe CUST_37231_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37231_PI426222305 | JHI_St_60k_v1 | DMT400068040 | GGAGCTAGAAGTCTGGGTTGAATTTGATCGAATTCAGTAACTTTTGCTCAAATAATGTAT |
All Microarray Probes Designed to Gene DMG400026461
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37231_PI426222305 | JHI_St_60k_v1 | DMT400068040 | GGAGCTAGAAGTCTGGGTTGAATTTGATCGAATTCAGTAACTTTTGCTCAAATAATGTAT |
Microarray Signals from CUST_37231_PI426222305
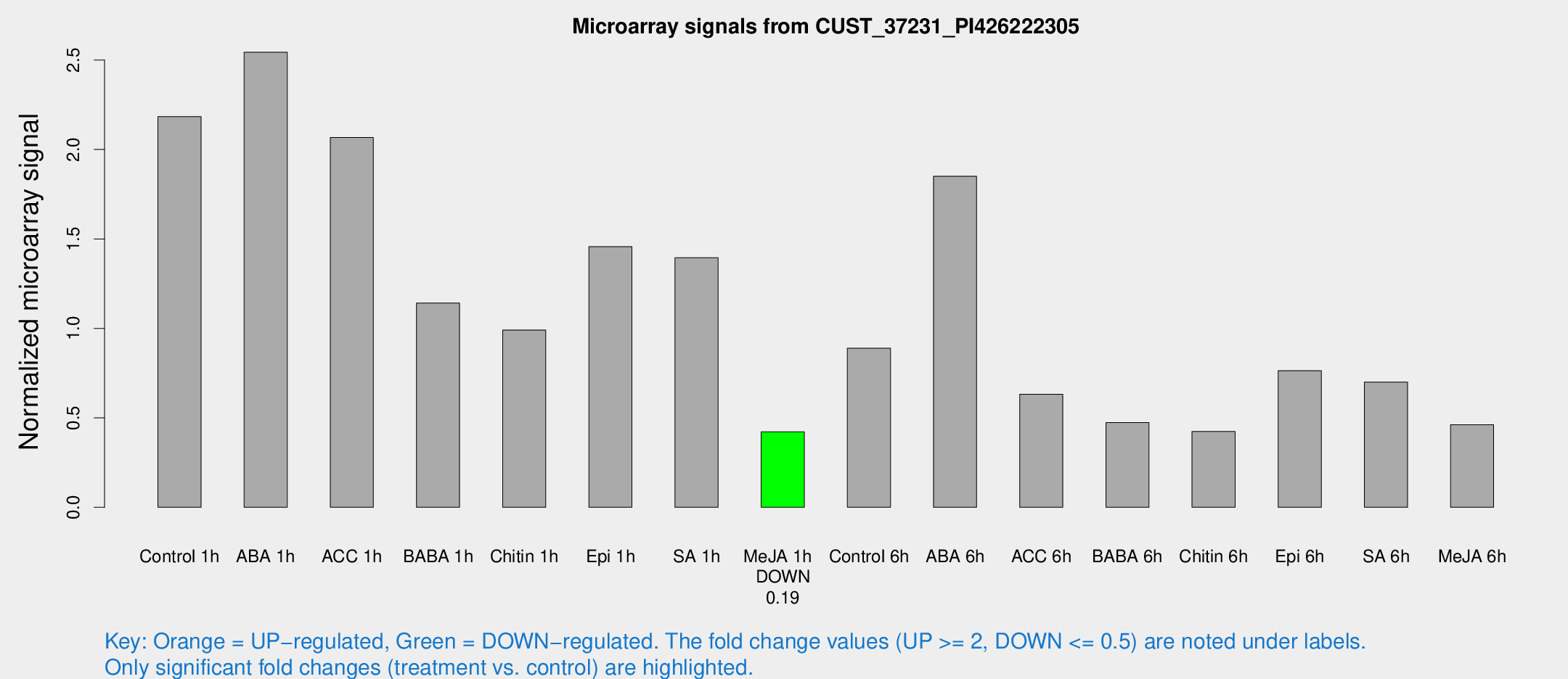
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 176.745 | 46.6769 | 2.1842 | 0.413587 |
| ABA 1h | 173.33 | 21.0373 | 2.54387 | 0.494669 |
| ACC 1h | 170.521 | 37.1955 | 2.0671 | 0.354411 |
| BABA 1h | 87.1444 | 18.0814 | 1.14231 | 0.15619 |
| Chitin 1h | 74.0387 | 23.2454 | 0.990858 | 0.338917 |
| Epi 1h | 96.3476 | 7.87429 | 1.45691 | 0.094658 |
| SA 1h | 112.052 | 17.7729 | 1.39572 | 0.179041 |
| Me-JA 1h | 26.1683 | 3.25801 | 0.421987 | 0.0526498 |
| Control 6h | 75.1112 | 20.7669 | 0.888898 | 0.234735 |
| ABA 6h | 153.98 | 25.1944 | 1.85073 | 0.243145 |
| ACC 6h | 58.7823 | 14.9991 | 0.632308 | 0.0840786 |
| BABA 6h | 41.4118 | 7.18156 | 0.473358 | 0.0727307 |
| Chitin 6h | 36.0194 | 7.30027 | 0.423884 | 0.117505 |
| Epi 6h | 68.6927 | 13.8792 | 0.763584 | 0.246015 |
| SA 6h | 54.1892 | 9.64713 | 0.700121 | 0.136997 |
| Me-JA 6h | 35.7384 | 5.58752 | 0.461971 | 0.0479622 |
Source Transcript PGSC0003DMT400068040 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G11590.1 | +1 | 2e-47 | 164 | 95/175 (54%) | Integrase-type DNA-binding superfamily protein | chr5:3727789-3728499 REVERSE LENGTH=236 |
