Probe CUST_36836_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36836_PI426222305 | JHI_St_60k_v1 | DMT400004170 | CCAACAAAGTGAACTTAGTTCAAGTGGCAACTAATGGAGCTATGAAGATTACCTAATTAT |
All Microarray Probes Designed to Gene DMG400001655
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36836_PI426222305 | JHI_St_60k_v1 | DMT400004170 | CCAACAAAGTGAACTTAGTTCAAGTGGCAACTAATGGAGCTATGAAGATTACCTAATTAT |
Microarray Signals from CUST_36836_PI426222305
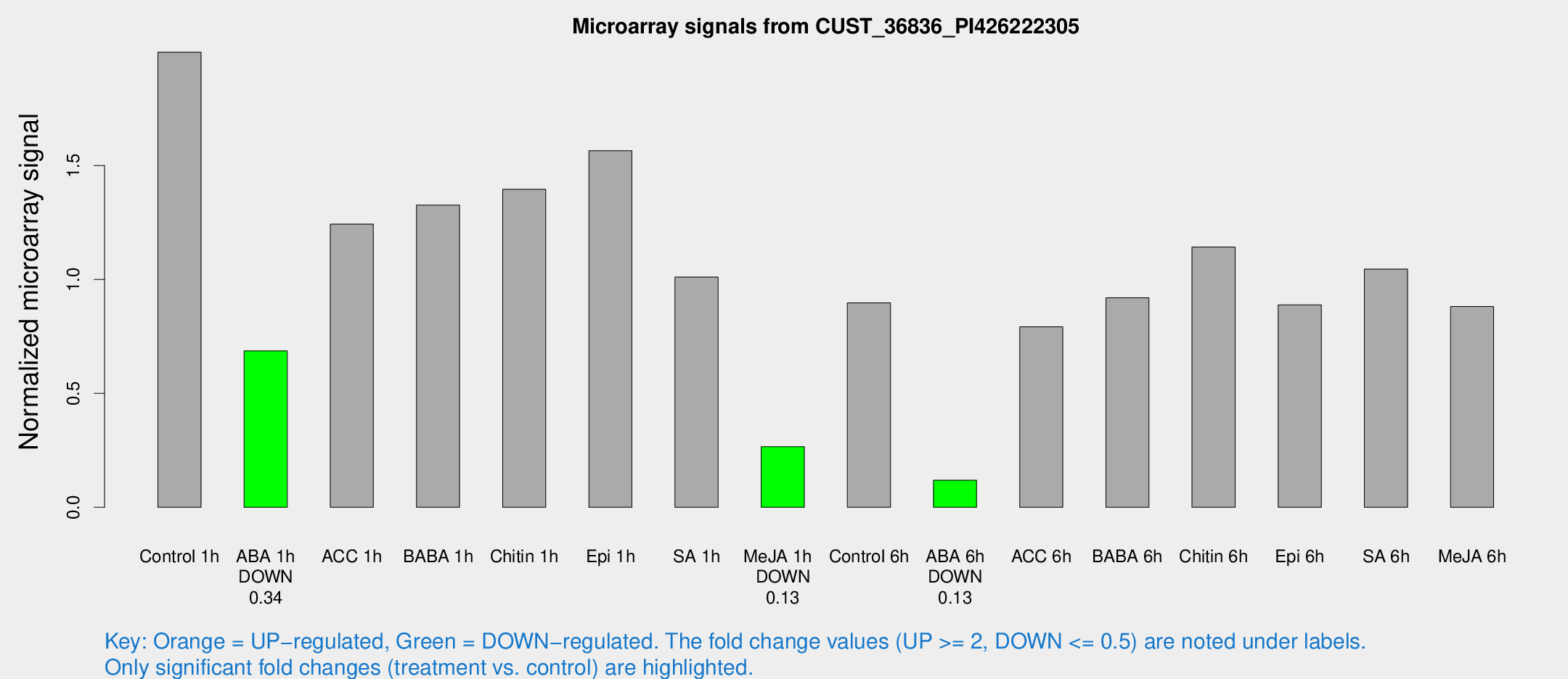
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 766.485 | 116.958 | 1.99738 | 0.222103 |
| ABA 1h | 227.848 | 14.3937 | 0.68646 | 0.0785337 |
| ACC 1h | 540.419 | 165.136 | 1.24303 | 0.396712 |
| BABA 1h | 510.886 | 118.241 | 1.3263 | 0.22581 |
| Chitin 1h | 476.045 | 55.896 | 1.39594 | 0.081324 |
| Epi 1h | 506.593 | 29.4822 | 1.56537 | 0.0910549 |
| SA 1h | 416.087 | 97.3439 | 1.01066 | 0.235518 |
| Me-JA 1h | 82.2784 | 10.0091 | 0.2659 | 0.0197641 |
| Control 6h | 366.085 | 102.718 | 0.897534 | 0.2101 |
| ABA 6h | 47.8652 | 5.88213 | 0.118487 | 0.0178614 |
| ACC 6h | 345.895 | 44.9522 | 0.792101 | 0.0704152 |
| BABA 6h | 403.283 | 91.8793 | 0.919478 | 0.197363 |
| Chitin 6h | 456.825 | 34.0491 | 1.14215 | 0.0668503 |
| Epi 6h | 378.991 | 40.5091 | 0.888731 | 0.0692829 |
| SA 6h | 393.962 | 52.831 | 1.0456 | 0.0614898 |
| Me-JA 6h | 344.746 | 80.721 | 0.881154 | 0.1467 |
Source Transcript PGSC0003DMT400004170 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G75590.1 | +1 | 3e-43 | 147 | 83/153 (54%) | SAUR-like auxin-responsive protein family | chr1:28383250-28383714 REVERSE LENGTH=154 |
