Probe CUST_36768_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36768_PI426222305 | JHI_St_60k_v1 | DMT400004086 | TTCTGCAGTAATGTGGAAGTACTGAGTTCAACTGAATGCAATAGAAACTGGATGTAAATT |
All Microarray Probes Designed to Gene DMG400001611
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36768_PI426222305 | JHI_St_60k_v1 | DMT400004086 | TTCTGCAGTAATGTGGAAGTACTGAGTTCAACTGAATGCAATAGAAACTGGATGTAAATT |
Microarray Signals from CUST_36768_PI426222305
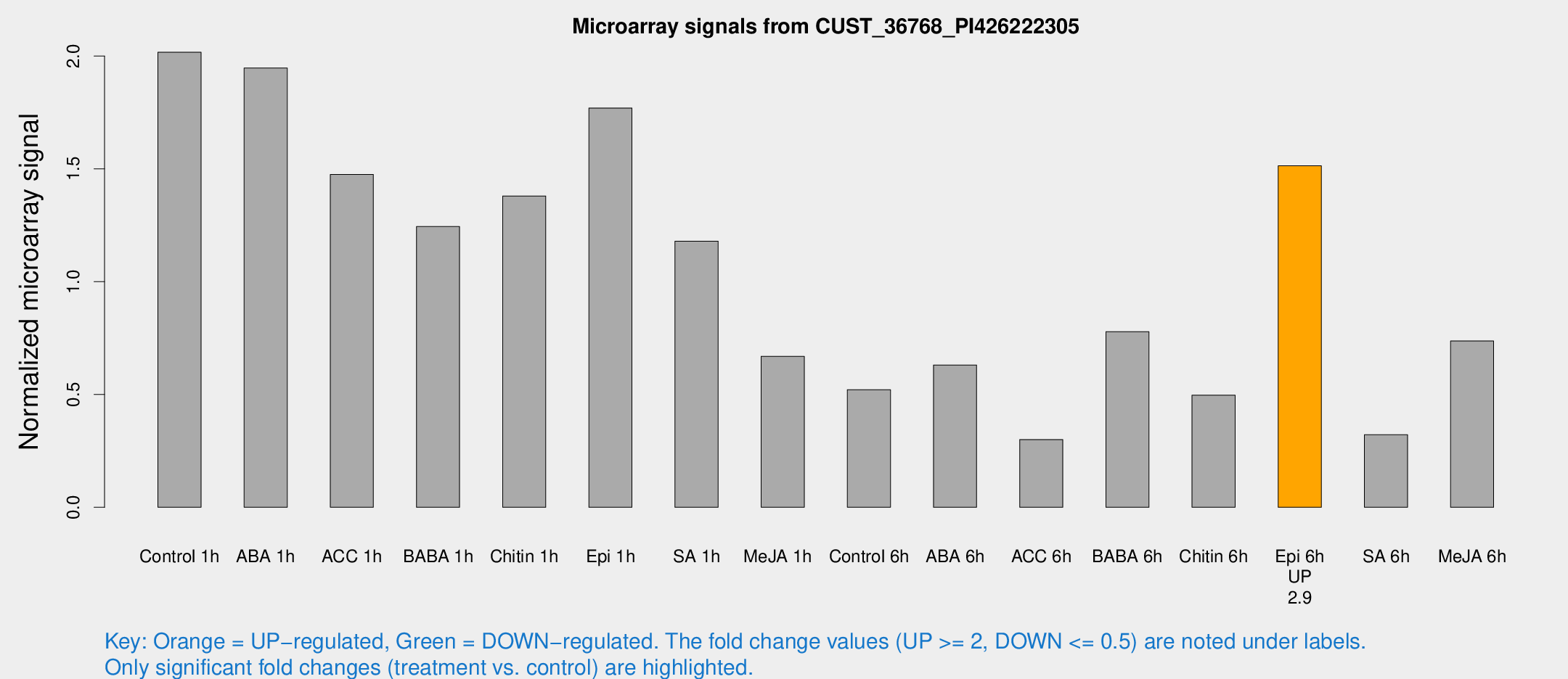
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.6679 | 6.09464 | 2.01703 | 0.221529 |
| ABA 1h | 32.5296 | 10.0336 | 1.94703 | 0.489004 |
| ACC 1h | 27.4054 | 6.57749 | 1.47545 | 0.267089 |
| BABA 1h | 23.0422 | 7.70941 | 1.2443 | 0.46772 |
| Chitin 1h | 21.4981 | 3.13824 | 1.37997 | 0.251281 |
| Epi 1h | 29.4391 | 9.56588 | 1.76978 | 0.621498 |
| SA 1h | 21.5704 | 4.25517 | 1.17983 | 0.187617 |
| Me-JA 1h | 9.91524 | 3.01229 | 0.66939 | 0.244278 |
| Control 6h | 9.6232 | 3.00953 | 0.521447 | 0.195929 |
| ABA 6h | 11.6447 | 3.15844 | 0.630766 | 0.17559 |
| ACC 6h | 6.03936 | 3.58448 | 0.300326 | 0.174258 |
| BABA 6h | 19.6235 | 9.25415 | 0.778434 | 0.488312 |
| Chitin 6h | 10.3074 | 3.79015 | 0.497346 | 0.210078 |
| Epi 6h | 29.8969 | 4.1993 | 1.51363 | 0.197519 |
| SA 6h | 5.45772 | 3.165 | 0.321417 | 0.186316 |
| Me-JA 6h | 13.5995 | 3.67824 | 0.737107 | 0.196638 |
Source Transcript PGSC0003DMT400004086 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G21210.1 | +1 | 1e-29 | 111 | 52/80 (65%) | SAUR-like auxin-responsive protein family | chr2:9085513-9085809 REVERSE LENGTH=98 |
