Probe CUST_36455_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36455_PI426222305 | JHI_St_60k_v1 | DMT400074570 | ACTGTCGAGAAGAGCCCTTGTGTATTTATTGTAACTGGGATGTTTAATTAGTGTTTTTCT |
All Microarray Probes Designed to Gene DMG400028989
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36455_PI426222305 | JHI_St_60k_v1 | DMT400074570 | ACTGTCGAGAAGAGCCCTTGTGTATTTATTGTAACTGGGATGTTTAATTAGTGTTTTTCT |
Microarray Signals from CUST_36455_PI426222305
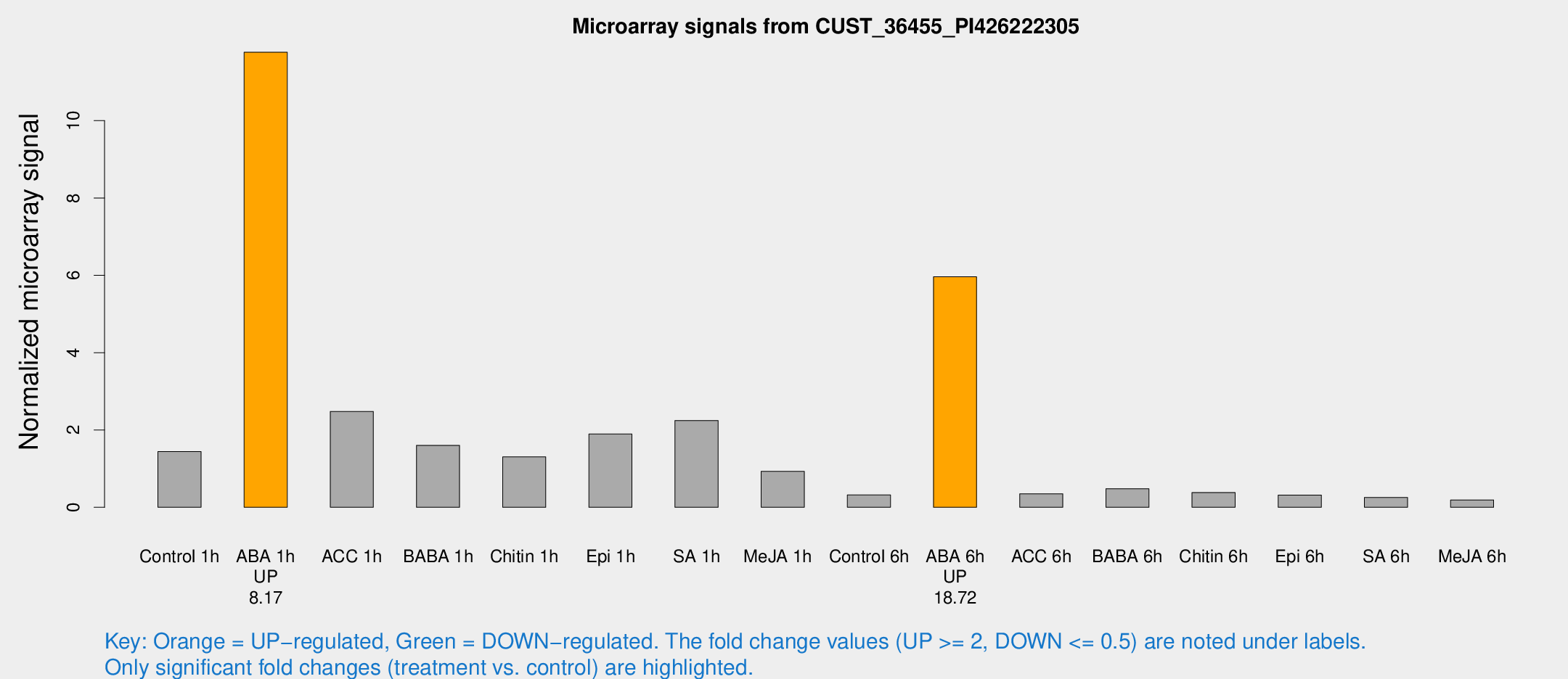
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 159.729 | 28.6598 | 1.44006 | 0.20238 |
| ABA 1h | 1168.59 | 242.419 | 11.7699 | 2.30267 |
| ACC 1h | 345.686 | 147.894 | 2.48028 | 1.29445 |
| BABA 1h | 166.234 | 13.0321 | 1.60309 | 0.0979116 |
| Chitin 1h | 136.443 | 39.4182 | 1.30476 | 0.285656 |
| Epi 1h | 177.468 | 18.675 | 1.89532 | 0.18538 |
| SA 1h | 247.136 | 14.6493 | 2.24381 | 0.26153 |
| Me-JA 1h | 83.0827 | 12.5311 | 0.929294 | 0.0690283 |
| Control 6h | 43.3043 | 17.2055 | 0.3184 | 0.168712 |
| ABA 6h | 680.108 | 42.684 | 5.96162 | 0.620455 |
| ACC 6h | 44.3518 | 7.94153 | 0.350522 | 0.0918972 |
| BABA 6h | 63.1542 | 18.0035 | 0.480728 | 0.161979 |
| Chitin 6h | 49.5634 | 18.4664 | 0.381946 | 0.155501 |
| Epi 6h | 38.4562 | 4.34689 | 0.317746 | 0.0360353 |
| SA 6h | 30.4194 | 9.04741 | 0.253535 | 0.0711193 |
| Me-JA 6h | 21.9803 | 6.50102 | 0.189519 | 0.0409275 |
Source Transcript PGSC0003DMT400074570 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G19875.1 | +3 | 8e-16 | 72 | 49/87 (56%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: response to oxidative stress; LOCATED IN: endomembrane system; EXPRESSED IN: 21 plant structures; EXPRESSED DURING: 13 growth stages; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT2G31940.1); Has 30201 Blast hits to 17322 proteins in 780 species: Archae - 12; Bacteria - 1396; Metazoa - 17338; Fungi - 3422; Plants - 5037; Viruses - 0; Other Eukaryotes - 2996 (source: NCBI BLink). | chr5:6718412-6718783 FORWARD LENGTH=123 |
