Probe CUST_36393_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36393_PI426222305 | JHI_St_60k_v1 | DMT400074549 | TGTTACACTTGCTTTAACACTCCCCTCTGTTTGTAAAGCCCCTGCTAACATCTCTGAATG |
All Microarray Probes Designed to Gene DMG400028977
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36393_PI426222305 | JHI_St_60k_v1 | DMT400074549 | TGTTACACTTGCTTTAACACTCCCCTCTGTTTGTAAAGCCCCTGCTAACATCTCTGAATG |
Microarray Signals from CUST_36393_PI426222305
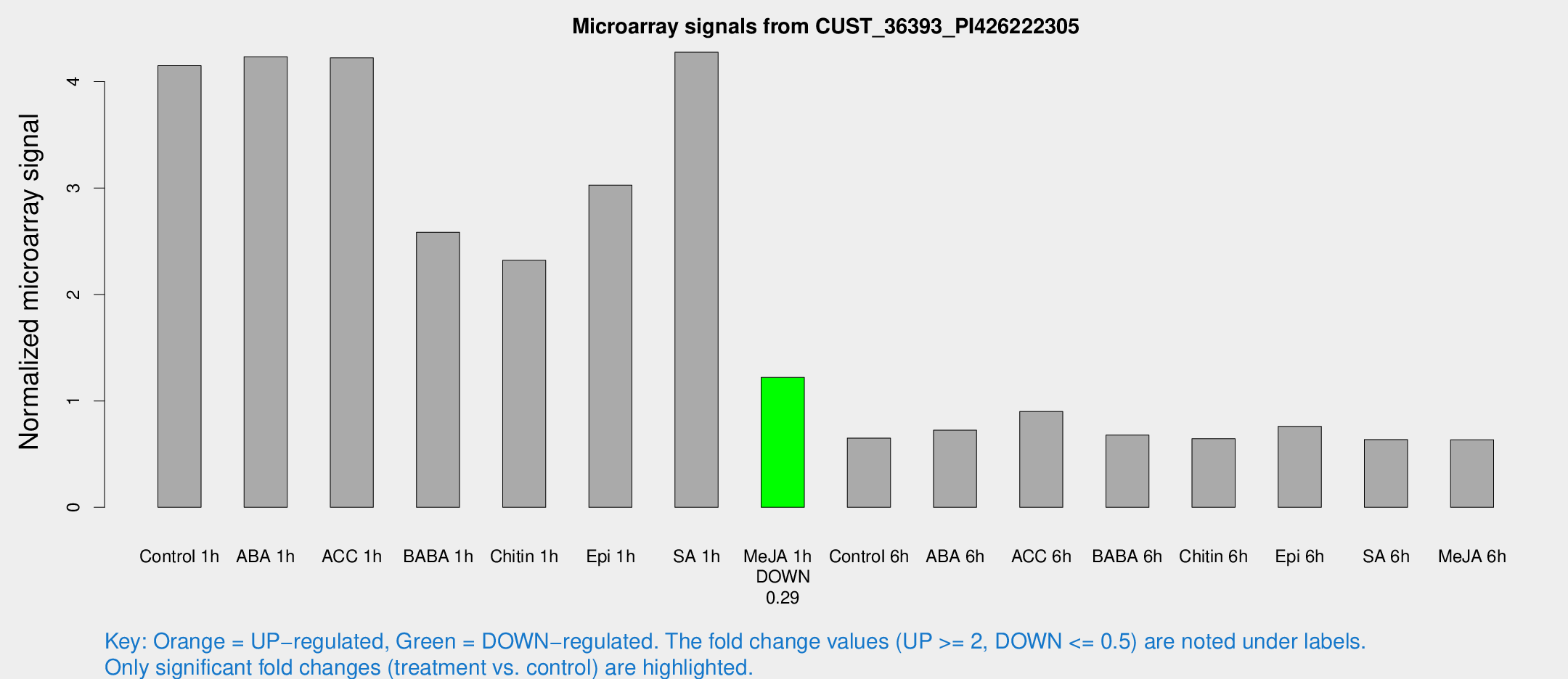
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4680.3 | 613.925 | 4.15101 | 0.290992 |
| ABA 1h | 4178.24 | 351.158 | 4.23477 | 0.583871 |
| ACC 1h | 5083.58 | 1202.11 | 4.2242 | 0.731504 |
| BABA 1h | 2871.9 | 537.334 | 2.58438 | 0.272945 |
| Chitin 1h | 2334.67 | 233.73 | 2.32113 | 0.223936 |
| Epi 1h | 2918.11 | 219.761 | 3.02778 | 0.262342 |
| SA 1h | 4912.95 | 497.619 | 4.27662 | 0.246931 |
| Me-JA 1h | 1112.81 | 104.791 | 1.22043 | 0.146388 |
| Control 6h | 760.407 | 157.548 | 0.650655 | 0.0974111 |
| ABA 6h | 854.744 | 49.6252 | 0.724166 | 0.0478716 |
| ACC 6h | 1155.35 | 121.596 | 0.89967 | 0.0586549 |
| BABA 6h | 842.928 | 48.9582 | 0.678406 | 0.0500875 |
| Chitin 6h | 760.065 | 44.132 | 0.643951 | 0.0375466 |
| Epi 6h | 958.047 | 89.465 | 0.760493 | 0.073839 |
| SA 6h | 736.324 | 162.024 | 0.637084 | 0.0837801 |
| Me-JA 6h | 718.054 | 111.291 | 0.634054 | 0.0528889 |
Source Transcript PGSC0003DMT400074549 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G44300.1 | +2 | 1e-51 | 171 | 76/111 (68%) | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | chr2:18307468-18308286 REVERSE LENGTH=204 |
