Probe CUST_36282_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36282_PI426222305 | JHI_St_60k_v1 | DMT400039832 | GATCCTCGACATCACTAGAGCATAAGCTGATTCAAAATTTCAAGATGATGGATGTATTAT |
All Microarray Probes Designed to Gene DMG400015401
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36282_PI426222305 | JHI_St_60k_v1 | DMT400039832 | GATCCTCGACATCACTAGAGCATAAGCTGATTCAAAATTTCAAGATGATGGATGTATTAT |
Microarray Signals from CUST_36282_PI426222305
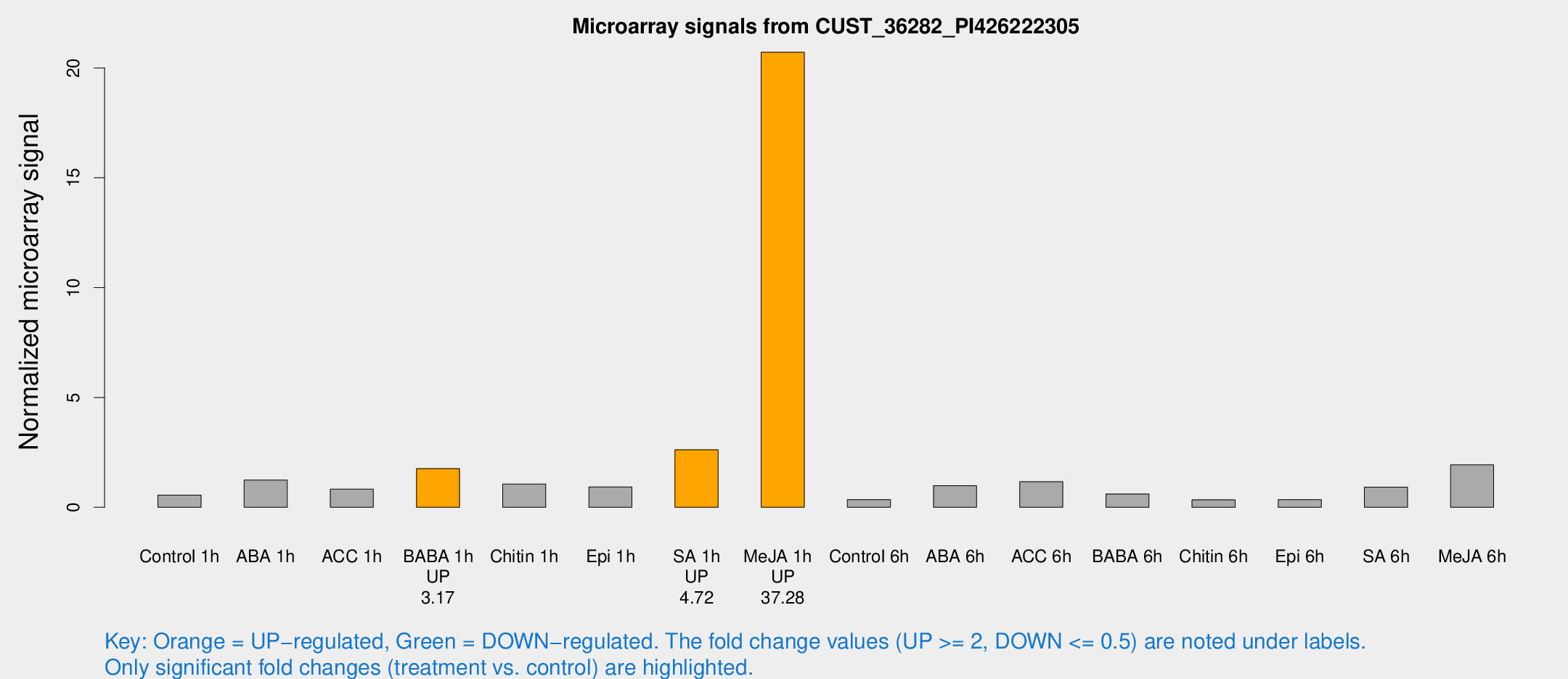
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 12.5048 | 3.23416 | 0.555648 | 0.16915 |
| ABA 1h | 26.0662 | 8.70043 | 1.24017 | 0.455962 |
| ACC 1h | 21.3953 | 8.84526 | 0.827714 | 0.37827 |
| BABA 1h | 39.6935 | 11.9161 | 1.76192 | 0.524006 |
| Chitin 1h | 19.7402 | 3.1845 | 1.0559 | 0.174193 |
| Epi 1h | 19.4479 | 6.5687 | 0.93111 | 0.446793 |
| SA 1h | 55.8628 | 4.72372 | 2.62213 | 0.210124 |
| Me-JA 1h | 368.413 | 78.8416 | 20.7169 | 3.76163 |
| Control 6h | 7.61526 | 3.07797 | 0.35325 | 0.157084 |
| ABA 6h | 22.892 | 5.09288 | 0.983368 | 0.279172 |
| ACC 6h | 29.013 | 7.13081 | 1.16305 | 0.409191 |
| BABA 6h | 20.9293 | 12.9583 | 0.609131 | 0.497251 |
| Chitin 6h | 7.94235 | 3.43931 | 0.342301 | 0.166918 |
| Epi 6h | 9.42676 | 3.80235 | 0.350526 | 0.167666 |
| SA 6h | 18.943 | 3.45958 | 0.915311 | 0.172714 |
| Me-JA 6h | 49.2271 | 17.7009 | 1.93161 | 1.54101 |
Source Transcript PGSC0003DMT400039832 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
