Probe CUST_36140_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36140_PI426222305 | JHI_St_60k_v1 | DMT400058532 | GATTTCTCACTTGTGGAACTAAGTATGTTGTTATTGTTGAATCAACAGCGATTGATACGA |
All Microarray Probes Designed to Gene DMG401022737
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36140_PI426222305 | JHI_St_60k_v1 | DMT400058532 | GATTTCTCACTTGTGGAACTAAGTATGTTGTTATTGTTGAATCAACAGCGATTGATACGA |
| CUST_36052_PI426222305 | JHI_St_60k_v1 | DMT400058531 | GATTTCTCACTTGTGGAACTAAGTATGTTGTTATTGTTGAATCAACAGCGATTGATACGA |
Microarray Signals from CUST_36140_PI426222305
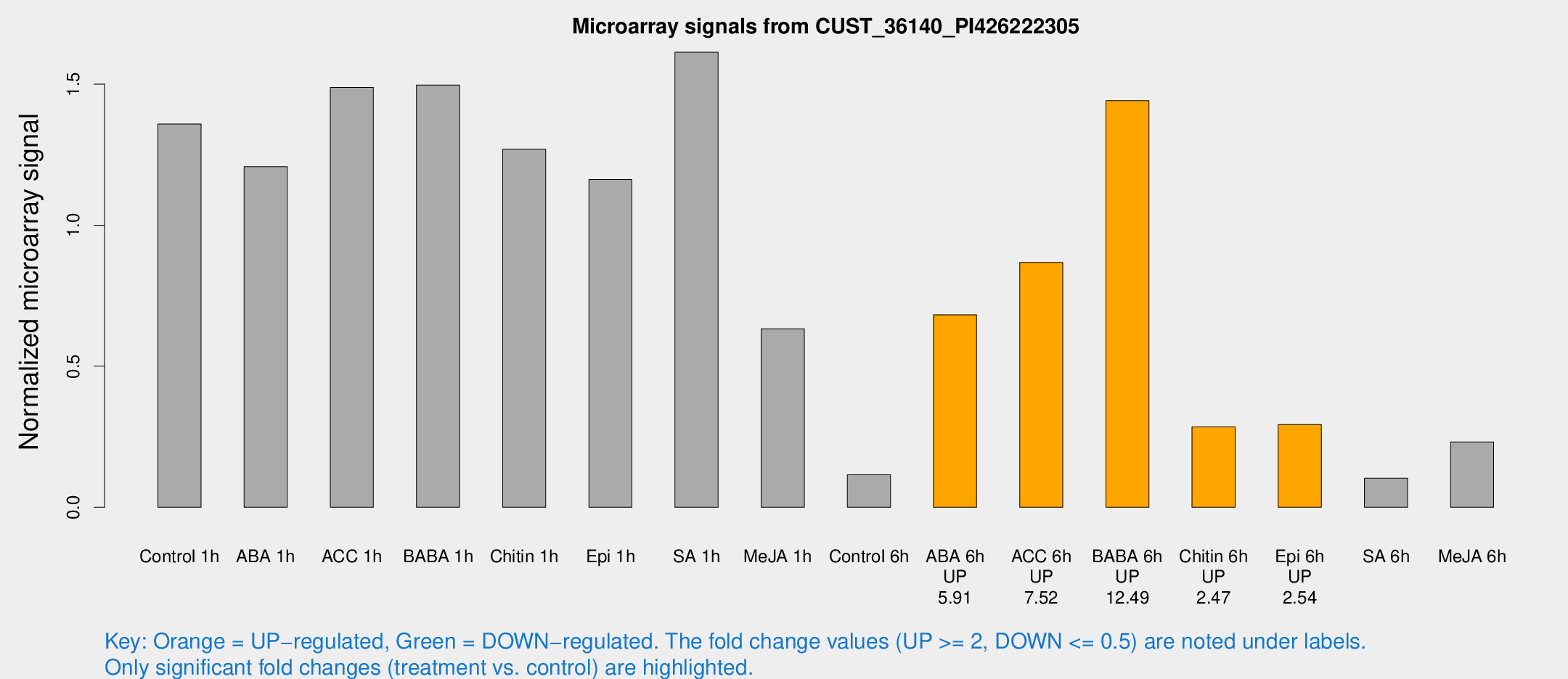
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 102.461 | 16.0422 | 1.35838 | 0.155998 |
| ABA 1h | 80.2056 | 10.8168 | 1.20719 | 0.087434 |
| ACC 1h | 126.285 | 36.5402 | 1.48801 | 0.449162 |
| BABA 1h | 116.16 | 30.8882 | 1.49682 | 0.325518 |
| Chitin 1h | 84.2061 | 5.72955 | 1.26944 | 0.142161 |
| Epi 1h | 74.1502 | 5.23512 | 1.16177 | 0.0821628 |
| SA 1h | 123.049 | 10.7264 | 1.61287 | 0.101451 |
| Me-JA 1h | 38.6837 | 4.6698 | 0.63289 | 0.0903912 |
| Control 6h | 9.29885 | 3.14728 | 0.115405 | 0.0474894 |
| ABA 6h | 58.9967 | 16.7521 | 0.682482 | 0.199886 |
| ACC 6h | 73.4478 | 5.66868 | 0.867771 | 0.139388 |
| BABA 6h | 141.283 | 61.1395 | 1.44105 | 0.607859 |
| Chitin 6h | 22.4458 | 3.64536 | 0.28501 | 0.0467975 |
| Epi 6h | 25.4731 | 5.50822 | 0.293299 | 0.0714375 |
| SA 6h | 8.15895 | 3.27695 | 0.102985 | 0.049151 |
| Me-JA 6h | 17.0727 | 3.21005 | 0.231583 | 0.0441277 |
Source Transcript PGSC0003DMT400058532 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G42010.1 | +3 | 0.0 | 1270 | 605/844 (72%) | phospholipase D beta 1 | chr2:17533018-17537990 REVERSE LENGTH=1083 |
