Probe CUST_36137_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36137_PI426222305 | JHI_St_60k_v1 | DMT400007898 | AGGAGCTCGGTTGGAAGGCAAAATATGGTATAAATGAGATGTGCAGGGACCAGTGGAAAT |
All Microarray Probes Designed to Gene DMG400003054
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36137_PI426222305 | JHI_St_60k_v1 | DMT400007898 | AGGAGCTCGGTTGGAAGGCAAAATATGGTATAAATGAGATGTGCAGGGACCAGTGGAAAT |
| CUST_36071_PI426222305 | JHI_St_60k_v1 | DMT400007897 | GCATAACTCTTTCATTTCACCAGTTGAAAGAGAAACTGATGGTGGGATCAAATCTTTTTT |
Microarray Signals from CUST_36137_PI426222305
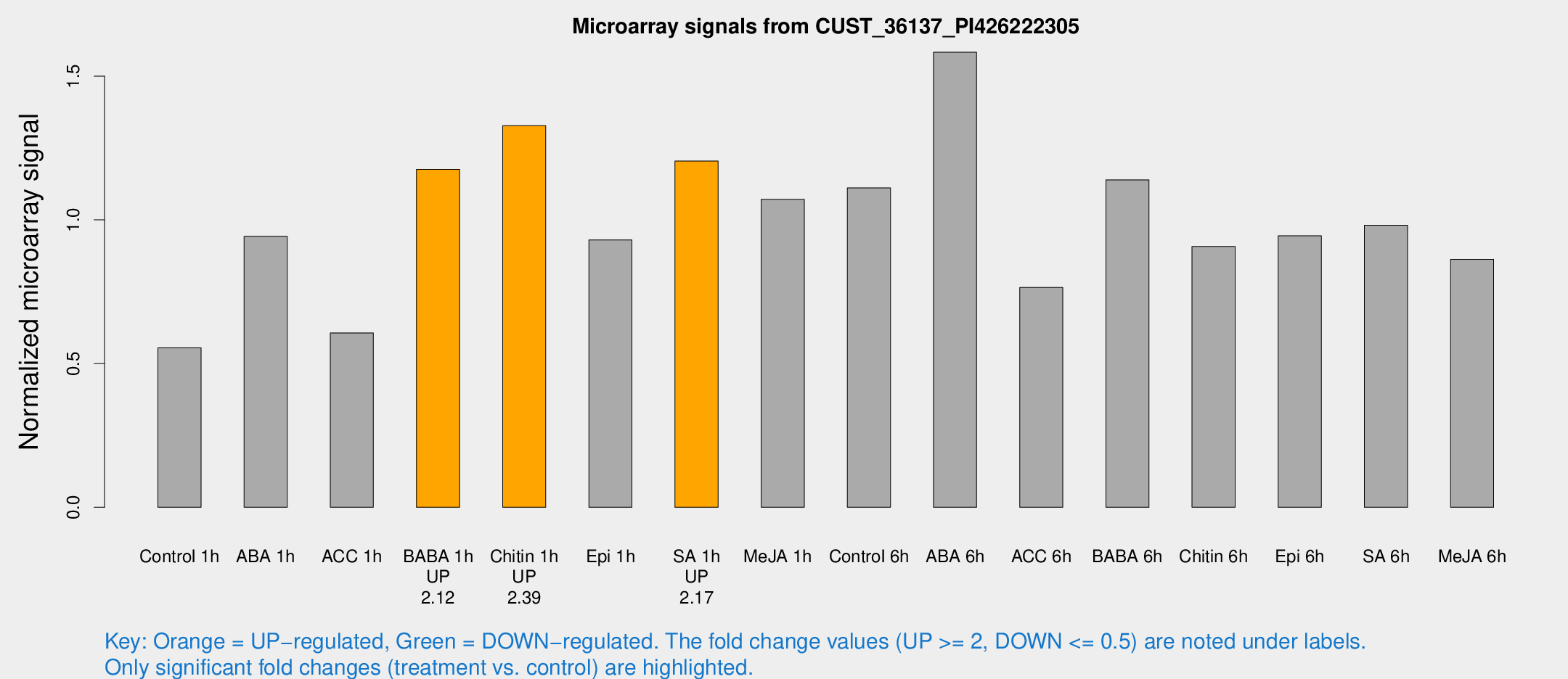
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1287.5 | 136.221 | 0.555109 | 0.0320899 |
| ABA 1h | 1995.43 | 376.704 | 0.942962 | 0.160218 |
| ACC 1h | 1556.86 | 417.179 | 0.606611 | 0.148419 |
| BABA 1h | 2703.18 | 491.94 | 1.1758 | 0.117594 |
| Chitin 1h | 2780.63 | 348.972 | 1.32772 | 0.125459 |
| Epi 1h | 1970.03 | 463.67 | 0.930171 | 0.23628 |
| SA 1h | 2905.92 | 462.638 | 1.20466 | 0.123185 |
| Me-JA 1h | 2067.66 | 359.46 | 1.07153 | 0.155742 |
| Control 6h | 2708.27 | 622.448 | 1.11122 | 0.194459 |
| ABA 6h | 3880.33 | 272.063 | 1.58322 | 0.16491 |
| ACC 6h | 2040.78 | 248.772 | 0.764697 | 0.044184 |
| BABA 6h | 2944.07 | 235.159 | 1.13933 | 0.0871837 |
| Chitin 6h | 2235.29 | 202.502 | 0.907481 | 0.066955 |
| Epi 6h | 2476.12 | 279.537 | 0.944474 | 0.0714178 |
| SA 6h | 2343.14 | 498.589 | 0.981392 | 0.121095 |
| Me-JA 6h | 2061.55 | 398.565 | 0.862773 | 0.127433 |
Source Transcript PGSC0003DMT400007898 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G12780.1 | +3 | 0.0 | 599 | 282/350 (81%) | UDP-D-glucose/UDP-D-galactose 4-epimerase 1 | chr1:4356124-4358120 REVERSE LENGTH=351 |
