Probe CUST_36083_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36083_PI426222305 | JHI_St_60k_v1 | DMT400007905 | AGCCACCTCTTTCTTGTTCAATTTGCATGGTTGAGTTTTGGTCATTAGTTGTCTTTTATA |
All Microarray Probes Designed to Gene DMG400003057
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36083_PI426222305 | JHI_St_60k_v1 | DMT400007905 | AGCCACCTCTTTCTTGTTCAATTTGCATGGTTGAGTTTTGGTCATTAGTTGTCTTTTATA |
Microarray Signals from CUST_36083_PI426222305
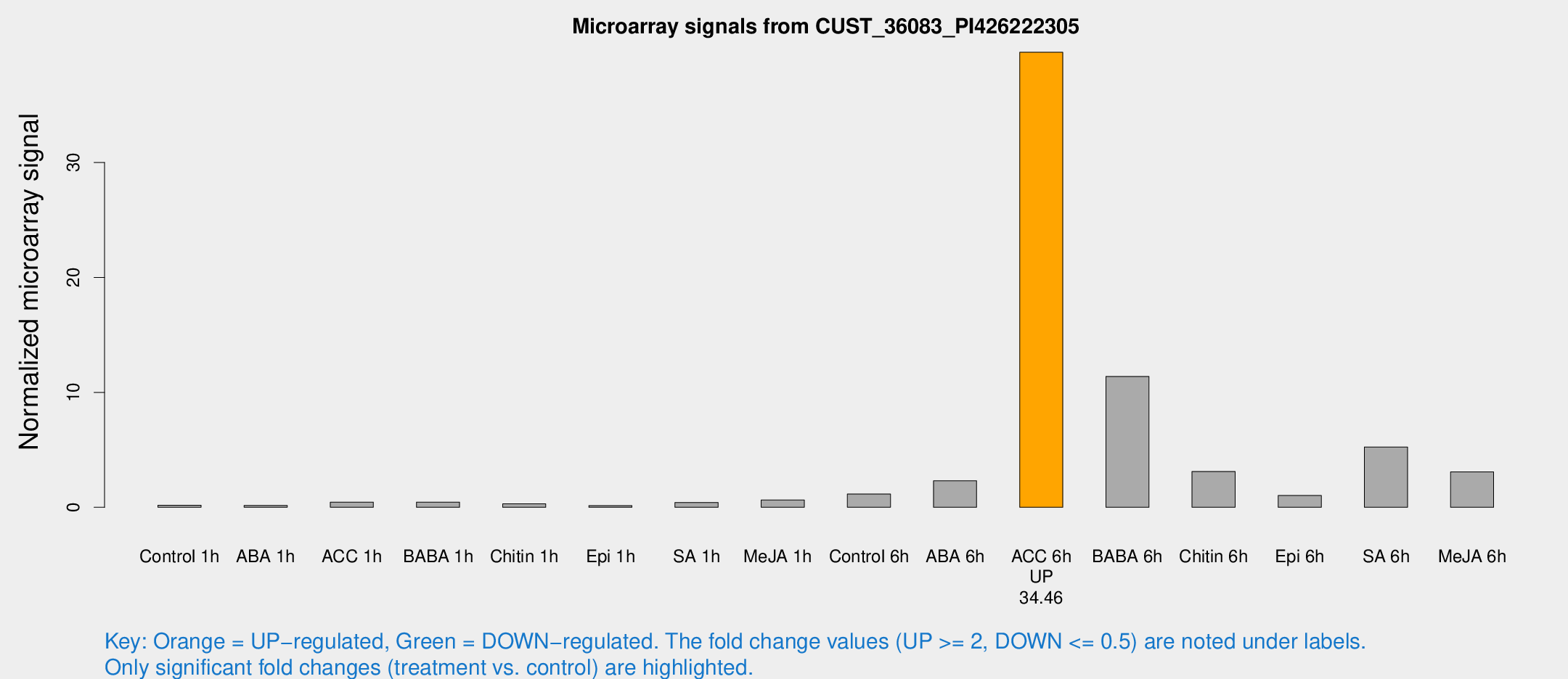
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.17387 | 3.72645 | 0.184084 | 0.0859439 |
| ABA 1h | 6.01213 | 3.03061 | 0.15534 | 0.081091 |
| ACC 1h | 20.0003 | 3.83558 | 0.448718 | 0.0873926 |
| BABA 1h | 25.3994 | 11.2695 | 0.453287 | 0.286434 |
| Chitin 1h | 14.2928 | 6.35544 | 0.300075 | 0.152135 |
| Epi 1h | 5.3631 | 3.11456 | 0.145771 | 0.0835143 |
| SA 1h | 26.6722 | 13.1069 | 0.422059 | 0.345603 |
| Me-JA 1h | 38.1203 | 26.5216 | 0.62867 | 0.710755 |
| Control 6h | 81.3188 | 45.5392 | 1.14924 | 1.2375 |
| ABA 6h | 127.564 | 58.562 | 2.3012 | 0.962931 |
| ACC 6h | 2103.2 | 515.712 | 39.5985 | 12.3041 |
| BABA 6h | 655.428 | 276.135 | 11.3887 | 5.35787 |
| Chitin 6h | 152.058 | 35.5039 | 3.11958 | 0.975731 |
| Epi 6h | 52.6817 | 11.844 | 1.03055 | 0.251671 |
| SA 6h | 260.078 | 84.2928 | 5.252 | 1.75408 |
| Me-JA 6h | 138.909 | 33.6185 | 3.08223 | 0.694981 |
Source Transcript PGSC0003DMT400007905 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G11650.1 | +1 | 3e-95 | 286 | 152/249 (61%) | osmotin 34 | chr4:7025127-7026113 REVERSE LENGTH=244 |
