Probe CUST_36052_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36052_PI426222305 | JHI_St_60k_v1 | DMT400058531 | GATTTCTCACTTGTGGAACTAAGTATGTTGTTATTGTTGAATCAACAGCGATTGATACGA |
All Microarray Probes Designed to Gene DMG401022737
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36140_PI426222305 | JHI_St_60k_v1 | DMT400058532 | GATTTCTCACTTGTGGAACTAAGTATGTTGTTATTGTTGAATCAACAGCGATTGATACGA |
| CUST_36052_PI426222305 | JHI_St_60k_v1 | DMT400058531 | GATTTCTCACTTGTGGAACTAAGTATGTTGTTATTGTTGAATCAACAGCGATTGATACGA |
Microarray Signals from CUST_36052_PI426222305
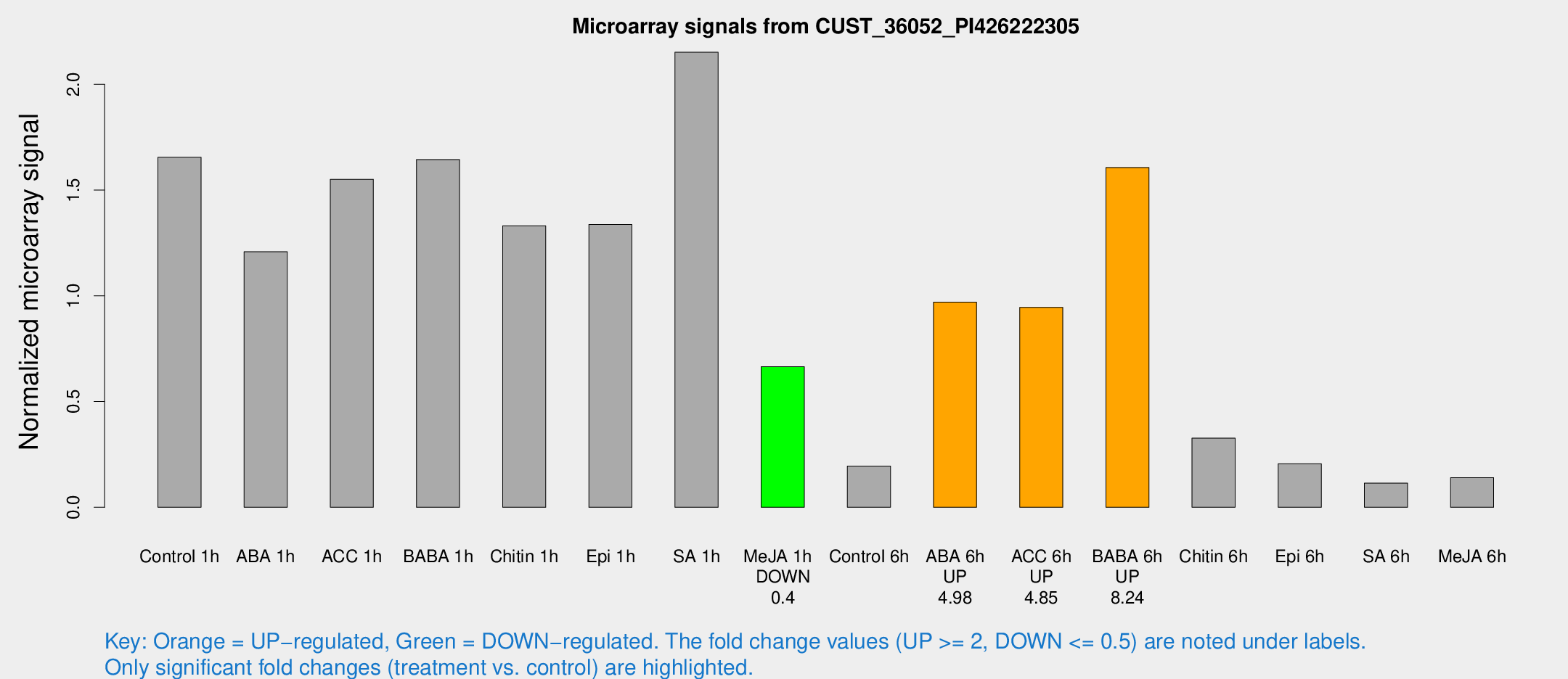
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 105.405 | 15.2559 | 1.6555 | 0.145642 |
| ABA 1h | 68.2814 | 9.85223 | 1.20868 | 0.10686 |
| ACC 1h | 113.795 | 34.5425 | 1.5511 | 0.537745 |
| BABA 1h | 106.341 | 25.2953 | 1.64476 | 0.297391 |
| Chitin 1h | 74.7795 | 5.49774 | 1.33116 | 0.130336 |
| Epi 1h | 74.4234 | 12.1071 | 1.33736 | 0.210713 |
| SA 1h | 138.682 | 9.38192 | 2.15211 | 0.135341 |
| Me-JA 1h | 34.4364 | 4.15383 | 0.66482 | 0.0990043 |
| Control 6h | 13.6383 | 4.24353 | 0.194916 | 0.0701424 |
| ABA 6h | 70.5245 | 20.1904 | 0.970033 | 0.259524 |
| ACC 6h | 67.9851 | 5.70741 | 0.945695 | 0.174485 |
| BABA 6h | 127.279 | 45.3219 | 1.60705 | 0.587566 |
| Chitin 6h | 22.1439 | 4.11132 | 0.327242 | 0.0639747 |
| Epi 6h | 15.9241 | 4.79139 | 0.206455 | 0.0756177 |
| SA 6h | 7.0676 | 3.77983 | 0.113947 | 0.061361 |
| Me-JA 6h | 9.40994 | 3.55664 | 0.140193 | 0.062176 |
Source Transcript PGSC0003DMT400058531 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G42010.1 | +3 | 0.0 | 1264 | 605/847 (71%) | phospholipase D beta 1 | chr2:17533018-17537990 REVERSE LENGTH=1083 |
