Probe CUST_36022_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36022_PI426222305 | JHI_St_60k_v1 | DMT400042829 | GAGAAGATCAGGATTCATGGAAAAAAGTGATGTTCTGGAAATGCTTCTCAATATTACTGA |
All Microarray Probes Designed to Gene DMG402016617
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36022_PI426222305 | JHI_St_60k_v1 | DMT400042829 | GAGAAGATCAGGATTCATGGAAAAAAGTGATGTTCTGGAAATGCTTCTCAATATTACTGA |
| CUST_36013_PI426222305 | JHI_St_60k_v1 | DMT400042827 | ACTAACTAACTCATTTAATTTGAGTCCCTTACTCACAGGTACTAGTTAATGTGTGGGCAA |
Microarray Signals from CUST_36022_PI426222305
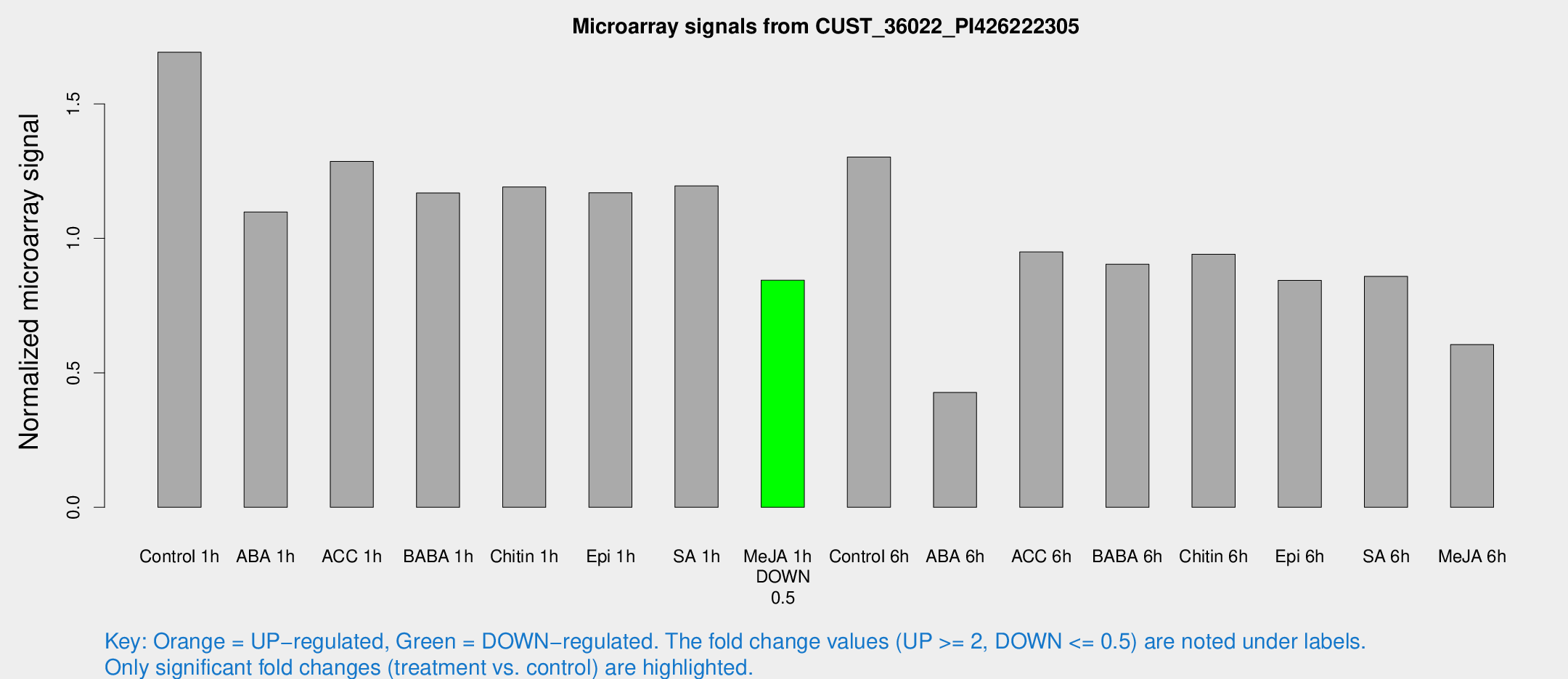
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 678.799 | 112.955 | 1.6921 | 0.163504 |
| ABA 1h | 381.998 | 30.7145 | 1.09815 | 0.0641589 |
| ACC 1h | 535.55 | 95.1503 | 1.28594 | 0.155381 |
| BABA 1h | 473.585 | 112.756 | 1.16879 | 0.21513 |
| Chitin 1h | 431.252 | 69.3593 | 1.1911 | 0.136596 |
| Epi 1h | 400.759 | 46.2535 | 1.16975 | 0.101315 |
| SA 1h | 491.73 | 73.9225 | 1.19523 | 0.119109 |
| Me-JA 1h | 271.491 | 24.1086 | 0.844362 | 0.0500269 |
| Control 6h | 587.915 | 183.25 | 1.3026 | 0.445961 |
| ABA 6h | 178.761 | 15.977 | 0.426919 | 0.0264544 |
| ACC 6h | 441.156 | 82.3209 | 0.949631 | 0.0769651 |
| BABA 6h | 405.182 | 63.4007 | 0.903908 | 0.107011 |
| Chitin 6h | 392.682 | 23.1548 | 0.940856 | 0.0553266 |
| Epi 6h | 382.25 | 64.1073 | 0.843622 | 0.178841 |
| SA 6h | 338.615 | 45.8526 | 0.858408 | 0.0507308 |
| Me-JA 6h | 256.664 | 70.1894 | 0.604883 | 0.137623 |
Source Transcript PGSC0003DMT400042829 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G61040.1 | +1 | 9e-105 | 236 | 139/311 (45%) | cytochrome P450, family 76, subfamily C, polypeptide 7 | chr3:22594074-22596125 REVERSE LENGTH=498 |
