Probe CUST_36013_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36013_PI426222305 | JHI_St_60k_v1 | DMT400042827 | ACTAACTAACTCATTTAATTTGAGTCCCTTACTCACAGGTACTAGTTAATGTGTGGGCAA |
All Microarray Probes Designed to Gene DMG402016617
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36013_PI426222305 | JHI_St_60k_v1 | DMT400042827 | ACTAACTAACTCATTTAATTTGAGTCCCTTACTCACAGGTACTAGTTAATGTGTGGGCAA |
| CUST_36022_PI426222305 | JHI_St_60k_v1 | DMT400042829 | GAGAAGATCAGGATTCATGGAAAAAAGTGATGTTCTGGAAATGCTTCTCAATATTACTGA |
Microarray Signals from CUST_36013_PI426222305
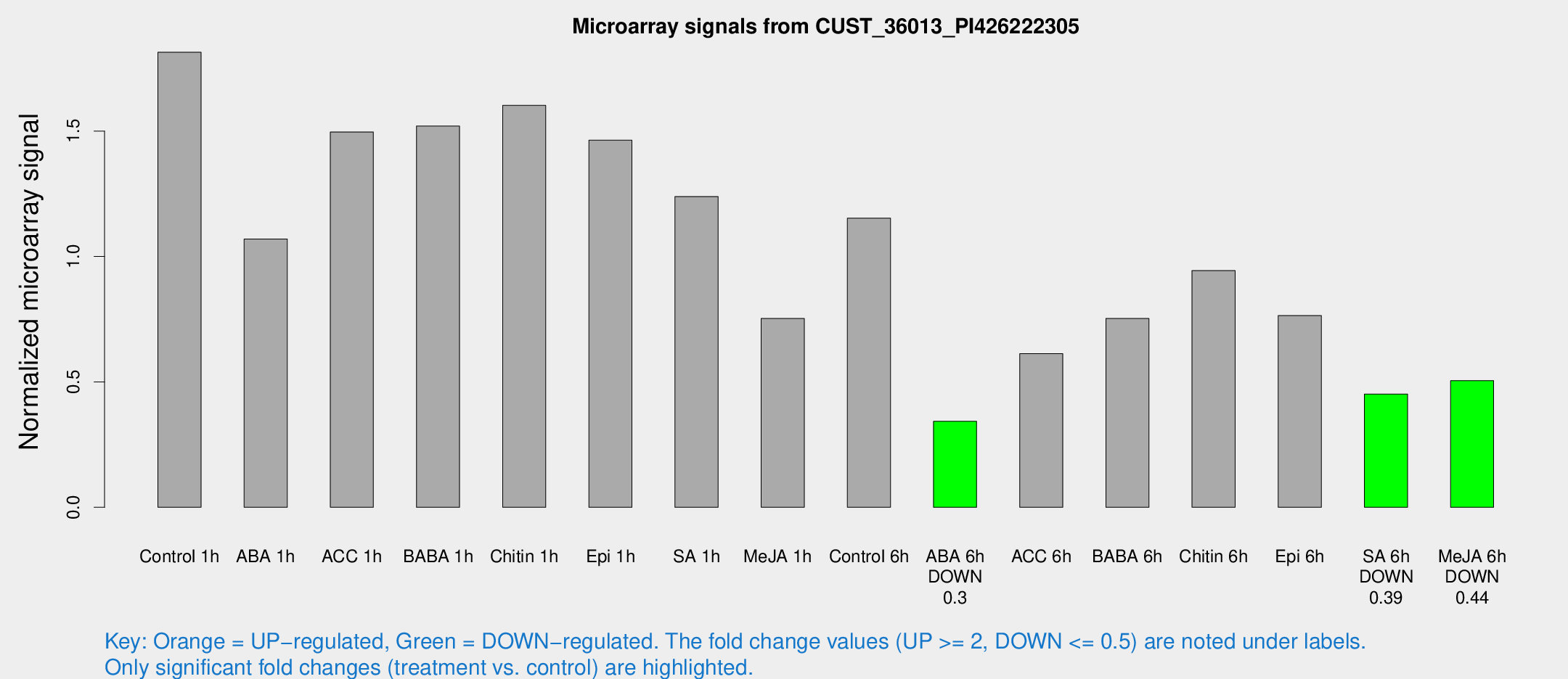
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 158.992 | 37.1064 | 1.81458 | 0.331225 |
| ABA 1h | 79.0462 | 5.48909 | 1.06971 | 0.0742016 |
| ACC 1h | 132.869 | 23.8522 | 1.49649 | 0.189382 |
| BABA 1h | 127.5 | 24.6919 | 1.52019 | 0.174173 |
| Chitin 1h | 121.577 | 14.0678 | 1.60278 | 0.1344 |
| Epi 1h | 109.816 | 20.4624 | 1.46372 | 0.295641 |
| SA 1h | 108.164 | 15.8914 | 1.23922 | 0.1091 |
| Me-JA 1h | 52.3091 | 8.04058 | 0.752686 | 0.0646084 |
| Control 6h | 103.58 | 24.8337 | 1.15295 | 0.226118 |
| ABA 6h | 30.4261 | 3.82286 | 0.342779 | 0.0434486 |
| ACC 6h | 61.0615 | 12.0655 | 0.612762 | 0.162031 |
| BABA 6h | 71.4371 | 9.06403 | 0.752961 | 0.0724784 |
| Chitin 6h | 83.8218 | 6.0681 | 0.943613 | 0.0682816 |
| Epi 6h | 74.2963 | 12.9808 | 0.764255 | 0.10603 |
| SA 6h | 40.8033 | 10.741 | 0.451503 | 0.0955652 |
| Me-JA 6h | 44.4282 | 10.1132 | 0.505103 | 0.085222 |
Source Transcript PGSC0003DMT400042827 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G45570.1 | +2 | 4e-40 | 144 | 65/106 (61%) | cytochrome P450, family 76, subfamily C, polypeptide 2 | chr2:18779935-18781922 REVERSE LENGTH=512 |
