Probe CUST_3578_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3578_PI426222305 | JHI_St_60k_v1 | DMT400064222 | CTTCTTTCAGAAATATCCTGATCGTGTCTTTCTTGCATGTTGTTAGTTTAGAAAGGTTTG |
All Microarray Probes Designed to Gene DMG400024951
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3569_PI426222305 | JHI_St_60k_v1 | DMT400064225 | ATATATATATGAGGTGCTATTTGCTCTTTAATGGTGAAAACATAGGCTGATGAACTACGG |
| CUST_3638_PI426222305 | JHI_St_60k_v1 | DMT400064219 | ATATATATATGAGGTGCTATTTGCTCTTTAATGGTGAAAACATAGGCTGATGAACTACGG |
| CUST_3744_PI426222305 | JHI_St_60k_v1 | DMT400064220 | AAGGGATTGTTTTGATCATGCTAAATGCGGGAACGGATACATCATCGGCGACAACAGAGT |
| CUST_3467_PI426222305 | JHI_St_60k_v1 | DMT400064224 | CGTTTGACCATACGATTTGGGACCCTAATTTCAAGTTTTAATTTGAAATCTGCACGAAAT |
| CUST_3578_PI426222305 | JHI_St_60k_v1 | DMT400064222 | CTTCTTTCAGAAATATCCTGATCGTGTCTTTCTTGCATGTTGTTAGTTTAGAAAGGTTTG |
| CUST_3628_PI426222305 | JHI_St_60k_v1 | DMT400064221 | CTCTCTAAACTCTATATATGAAGAGGAAAAAGTAGAGATGACCATCTGCATTTACACTCT |
Microarray Signals from CUST_3578_PI426222305
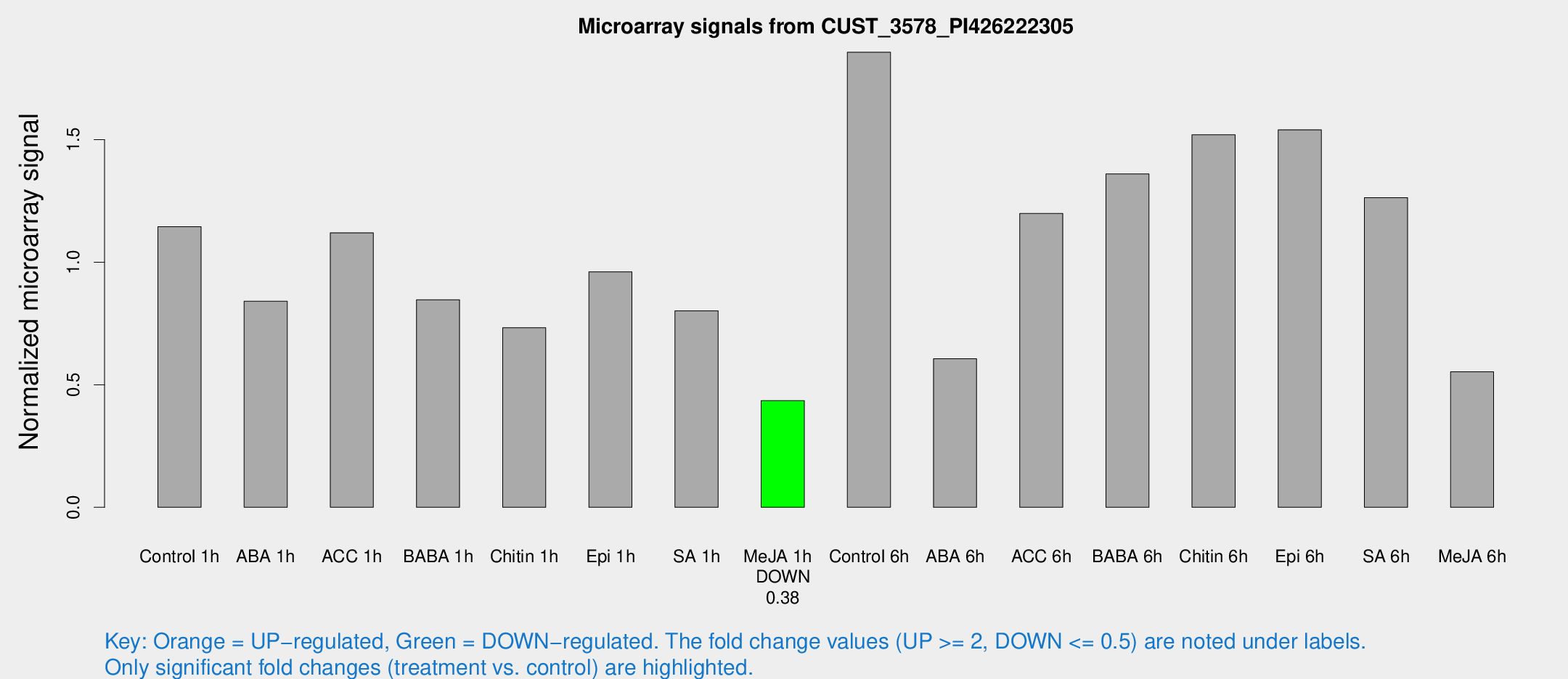
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2012.7 | 387.963 | 1.14517 | 0.143681 |
| ABA 1h | 1274.72 | 139.846 | 0.840559 | 0.0663222 |
| ACC 1h | 1974.85 | 231.87 | 1.12017 | 0.0869579 |
| BABA 1h | 1429.7 | 242.137 | 0.846829 | 0.0714497 |
| Chitin 1h | 1142.82 | 181.068 | 0.732638 | 0.1372 |
| Epi 1h | 1421.87 | 137.395 | 0.960862 | 0.0681892 |
| SA 1h | 1470.78 | 305.743 | 0.801965 | 0.201925 |
| Me-JA 1h | 604.18 | 36.8121 | 0.435183 | 0.0262061 |
| Control 6h | 3312.68 | 666.004 | 1.85684 | 0.257752 |
| ABA 6h | 1108.77 | 134.722 | 0.606299 | 0.0955001 |
| ACC 6h | 2418.6 | 459.544 | 1.19901 | 0.102328 |
| BABA 6h | 2638.03 | 395.262 | 1.36026 | 0.176359 |
| Chitin 6h | 2772.85 | 305.035 | 1.51983 | 0.117194 |
| Epi 6h | 2996.59 | 400.254 | 1.5403 | 0.286228 |
| SA 6h | 2178.54 | 343.554 | 1.26305 | 0.0729564 |
| Me-JA 6h | 1166.15 | 446.064 | 0.553207 | 0.281528 |
Source Transcript PGSC0003DMT400064222 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G37360.1 | +2 | 8e-74 | 183 | 85/119 (71%) | cytochrome P450, family 81, subfamily D, polypeptide 2 | chr4:17567124-17568858 REVERSE LENGTH=499 |
