Probe CUST_35662_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35662_PI426222305 | JHI_St_60k_v1 | DMT400087482 | GAGGTTCAAGATCTATCATTGGATGAGTTGCGGTGTCAGAGAATTCAAAGATATGGTAGA |
All Microarray Probes Designed to Gene DMG400037053
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35662_PI426222305 | JHI_St_60k_v1 | DMT400087482 | GAGGTTCAAGATCTATCATTGGATGAGTTGCGGTGTCAGAGAATTCAAAGATATGGTAGA |
Microarray Signals from CUST_35662_PI426222305
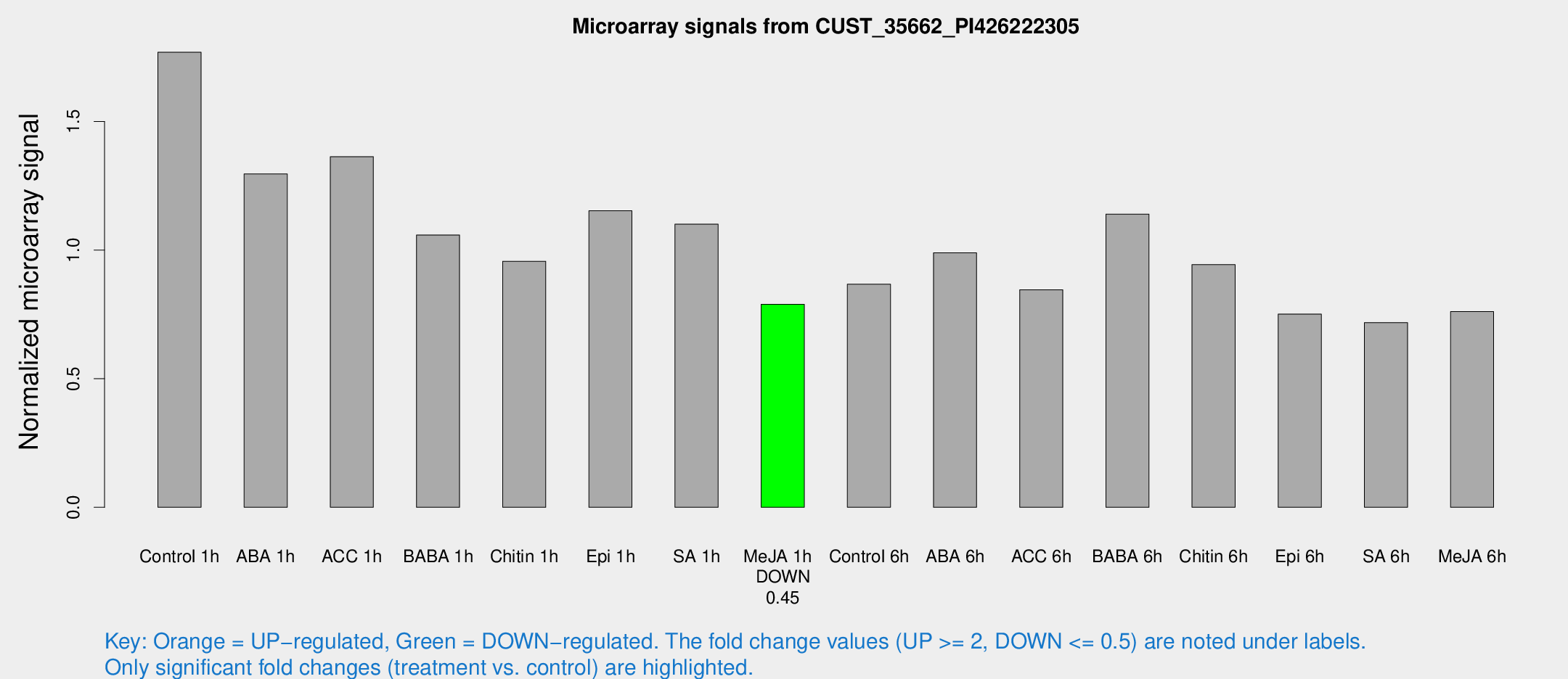
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 128.437 | 14.7236 | 1.76956 | 0.136604 |
| ABA 1h | 82.7751 | 7.6917 | 1.29608 | 0.0951 |
| ACC 1h | 101.581 | 12.3256 | 1.36317 | 0.12344 |
| BABA 1h | 79.7505 | 21.8556 | 1.05879 | 0.22996 |
| Chitin 1h | 61.6569 | 5.17478 | 0.956618 | 0.0801338 |
| Epi 1h | 71.6133 | 5.51689 | 1.15289 | 0.0893751 |
| SA 1h | 82.5518 | 11.5688 | 1.10105 | 0.0878982 |
| Me-JA 1h | 46.3918 | 4.74131 | 0.789133 | 0.0808896 |
| Control 6h | 70.592 | 21.7963 | 0.867387 | 0.270699 |
| ABA 6h | 76.6462 | 9.89913 | 0.989297 | 0.103458 |
| ACC 6h | 70.1546 | 7.0964 | 0.845907 | 0.0756669 |
| BABA 6h | 92.9736 | 12.2328 | 1.1401 | 0.124722 |
| Chitin 6h | 72.5308 | 6.82133 | 0.943618 | 0.104657 |
| Epi 6h | 67.6614 | 23.6309 | 0.75136 | 0.25645 |
| SA 6h | 54.1642 | 12.1761 | 0.718248 | 0.106636 |
| Me-JA 6h | 60.6604 | 17.9285 | 0.760996 | 0.218878 |
Source Transcript PGSC0003DMT400087482 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G17611.1 | +1 | 3e-44 | 151 | 91/172 (53%) | RHOMBOID-like protein 14 | chr3:6024946-6026173 FORWARD LENGTH=334 |
