Probe CUST_35389_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35389_PI426222305 | JHI_St_60k_v1 | DMT400079445 | CTTTGGGGGGTTTGTGTTGAAGTGAAGCAACAACCATGTCTACTCTTGTTAATTACAGTG |
All Microarray Probes Designed to Gene DMG400030928
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35354_PI426222305 | JHI_St_60k_v1 | DMT400079444 | CGATTGTACAGGTTCCTTGTCTTTCAGATTTGTACTTTGCAACTACTTTGTTATGTGTAA |
| CUST_35389_PI426222305 | JHI_St_60k_v1 | DMT400079445 | CTTTGGGGGGTTTGTGTTGAAGTGAAGCAACAACCATGTCTACTCTTGTTAATTACAGTG |
Microarray Signals from CUST_35389_PI426222305
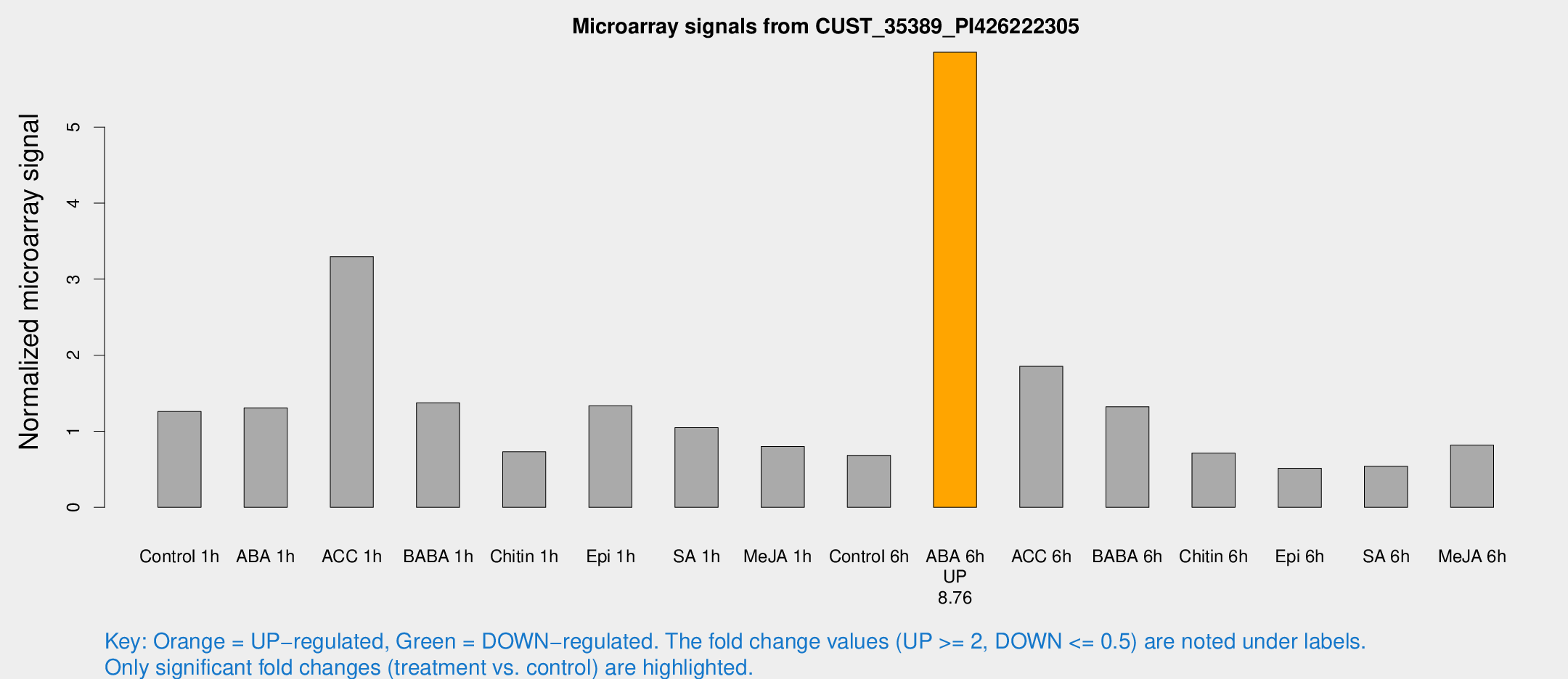
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 49.7304 | 4.32941 | 1.25977 | 0.14459 |
| ABA 1h | 51.7379 | 17.2196 | 1.30749 | 0.487668 |
| ACC 1h | 153.866 | 56.1681 | 3.29769 | 1.1779 |
| BABA 1h | 55.3179 | 12.4428 | 1.3733 | 0.468338 |
| Chitin 1h | 25.8783 | 3.52938 | 0.73052 | 0.101347 |
| Epi 1h | 46.7915 | 7.9094 | 1.33461 | 0.279728 |
| SA 1h | 42.4279 | 4.01572 | 1.04794 | 0.125519 |
| Me-JA 1h | 26.5312 | 4.67509 | 0.799537 | 0.152002 |
| Control 6h | 27.4154 | 3.92861 | 0.68308 | 0.111858 |
| ABA 6h | 254.691 | 33.5343 | 5.9843 | 0.639587 |
| ACC 6h | 96.9358 | 32.7379 | 1.85435 | 0.704007 |
| BABA 6h | 64.3914 | 21.6593 | 1.32267 | 0.396228 |
| Chitin 6h | 31.5007 | 6.56842 | 0.714334 | 0.204475 |
| Epi 6h | 26.2713 | 9.35083 | 0.513749 | 0.258292 |
| SA 6h | 24.2468 | 9.42509 | 0.540759 | 0.219339 |
| Me-JA 6h | 38.6952 | 13.5423 | 0.818712 | 0.370505 |
Source Transcript PGSC0003DMT400079445 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G25490.1 | +1 | 0.0 | 627 | 337/647 (52%) | EIN3-binding F box protein 1 | chr2:10848018-10850275 REVERSE LENGTH=628 |
