Probe CUST_3537_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3537_PI426222305 | JHI_St_60k_v1 | DMT400064187 | GTGTTGTGTTTGCTACTGCTTGGGTTCCTCTTTAGTTAATTAATTTCATTTGACTTAGCT |
All Microarray Probes Designed to Gene DMG400024937
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3537_PI426222305 | JHI_St_60k_v1 | DMT400064187 | GTGTTGTGTTTGCTACTGCTTGGGTTCCTCTTTAGTTAATTAATTTCATTTGACTTAGCT |
Microarray Signals from CUST_3537_PI426222305
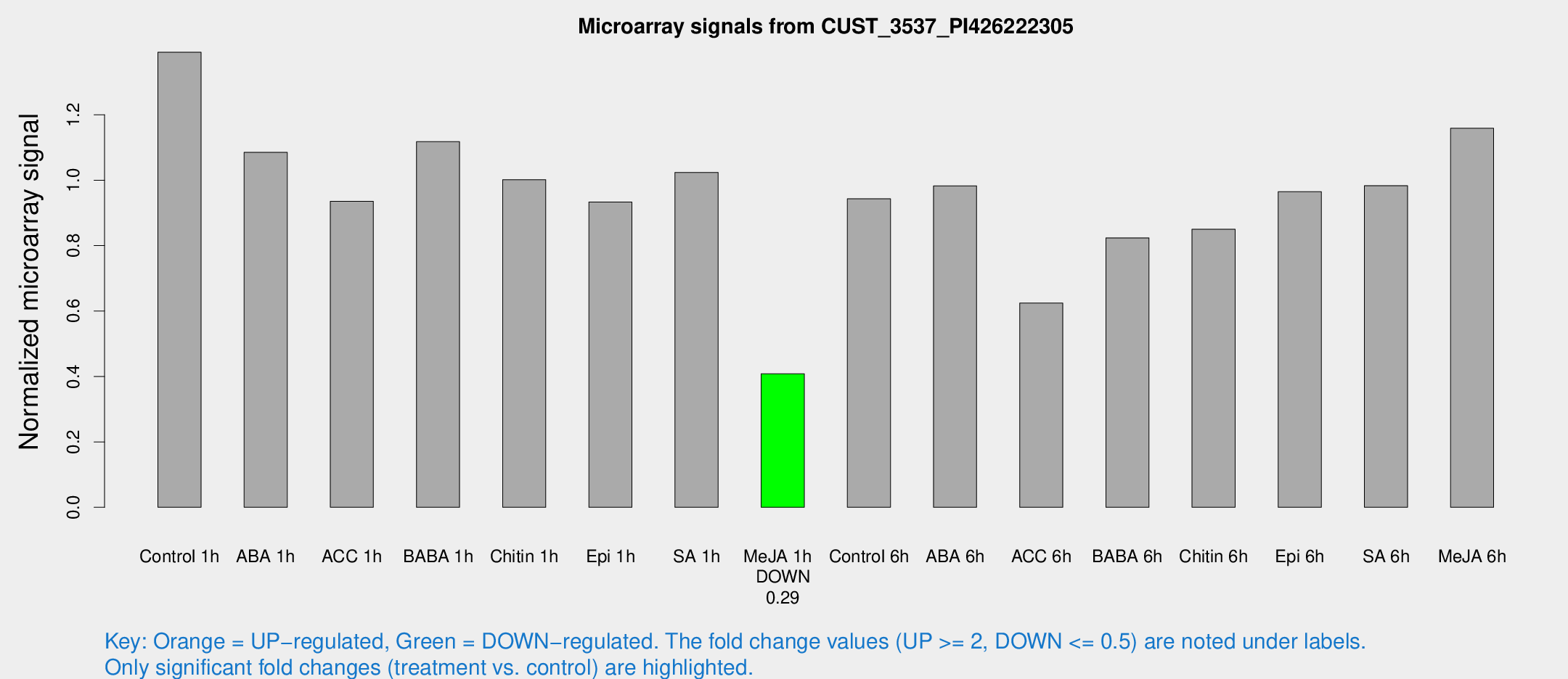
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3452.33 | 209.418 | 1.39115 | 0.0803309 |
| ABA 1h | 2586.11 | 742.609 | 1.08509 | 0.30218 |
| ACC 1h | 2468.42 | 506.293 | 0.935173 | 0.140843 |
| BABA 1h | 2780.4 | 552.888 | 1.11772 | 0.133057 |
| Chitin 1h | 2230.56 | 129.04 | 1.0016 | 0.0947432 |
| Epi 1h | 2025.08 | 230.832 | 0.933338 | 0.0918194 |
| SA 1h | 2612.64 | 178.893 | 1.02354 | 0.089192 |
| Me-JA 1h | 823.901 | 47.721 | 0.408031 | 0.0412532 |
| Control 6h | 2502.73 | 600.859 | 0.943047 | 0.170744 |
| ABA 6h | 2592.46 | 150.689 | 0.982701 | 0.0728933 |
| ACC 6h | 1794.83 | 203.831 | 0.624228 | 0.0676724 |
| BABA 6h | 2309.23 | 265.437 | 0.823698 | 0.0813236 |
| Chitin 6h | 2314.91 | 404.271 | 0.849809 | 0.189644 |
| Epi 6h | 2867.1 | 645.372 | 0.964559 | 0.348607 |
| SA 6h | 2479.45 | 414.719 | 0.983257 | 0.0757053 |
| Me-JA 6h | 2911.43 | 397.368 | 1.15861 | 0.0727774 |
Source Transcript PGSC0003DMT400064187 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G49950.1 | +3 | 2e-172 | 500 | 252/406 (62%) | GRAS family transcription factor | chr3:18522570-18523802 FORWARD LENGTH=410 |
