Probe CUST_35354_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35354_PI426222305 | JHI_St_60k_v1 | DMT400079444 | CGATTGTACAGGTTCCTTGTCTTTCAGATTTGTACTTTGCAACTACTTTGTTATGTGTAA |
All Microarray Probes Designed to Gene DMG400030928
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35389_PI426222305 | JHI_St_60k_v1 | DMT400079445 | CTTTGGGGGGTTTGTGTTGAAGTGAAGCAACAACCATGTCTACTCTTGTTAATTACAGTG |
| CUST_35354_PI426222305 | JHI_St_60k_v1 | DMT400079444 | CGATTGTACAGGTTCCTTGTCTTTCAGATTTGTACTTTGCAACTACTTTGTTATGTGTAA |
Microarray Signals from CUST_35354_PI426222305
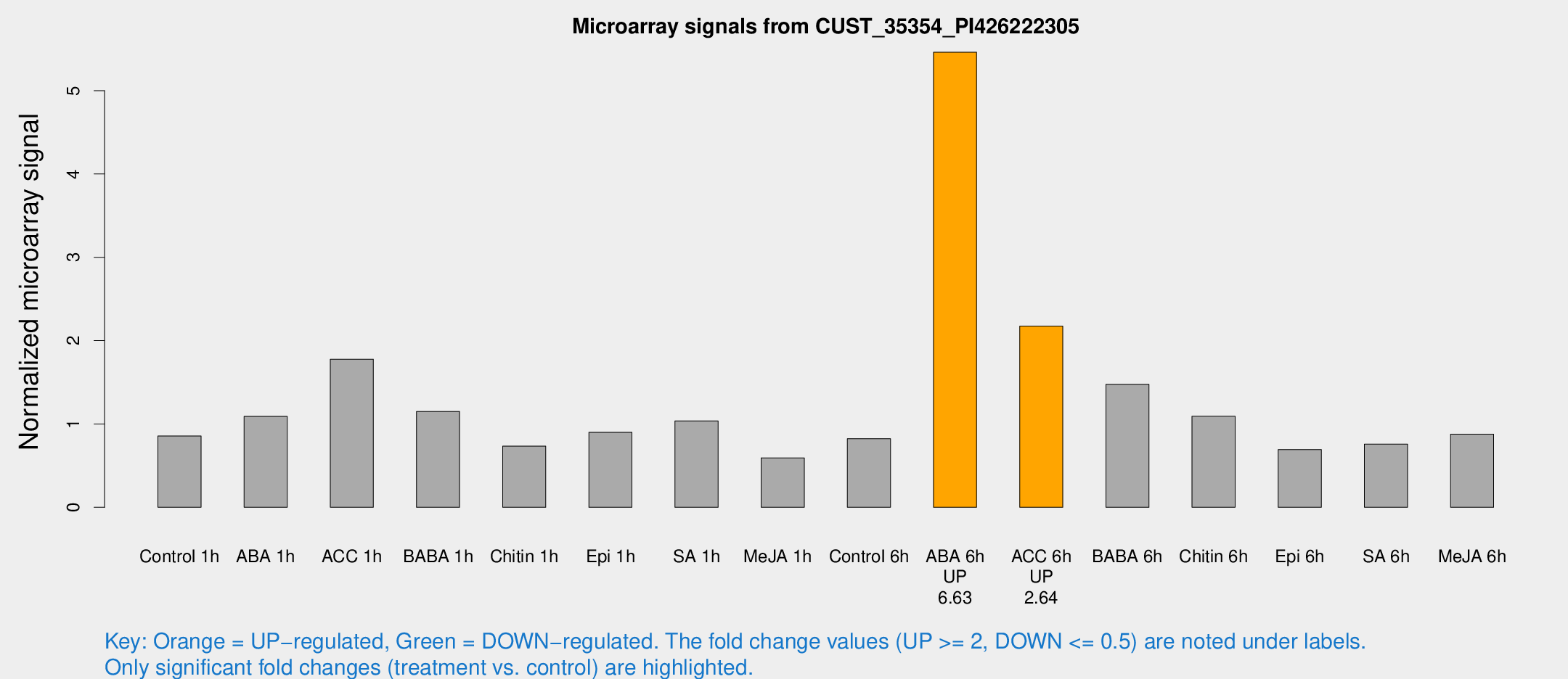
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3068.06 | 212.466 | 0.855003 | 0.0493733 |
| ABA 1h | 3737.76 | 983.483 | 1.09159 | 0.29516 |
| ACC 1h | 7682.99 | 2672.36 | 1.77624 | 0.715853 |
| BABA 1h | 4079.1 | 686.912 | 1.15018 | 0.102168 |
| Chitin 1h | 2380.76 | 245.039 | 0.733667 | 0.0423731 |
| Epi 1h | 2885.04 | 519.16 | 0.900268 | 0.171708 |
| SA 1h | 3828.98 | 287.02 | 1.03734 | 0.0598989 |
| Me-JA 1h | 1756.82 | 208.551 | 0.593639 | 0.034297 |
| Control 6h | 3095.25 | 622.286 | 0.823517 | 0.116231 |
| ABA 6h | 20843 | 1464.82 | 5.46146 | 0.315319 |
| ACC 6h | 9047.17 | 1142.08 | 2.17471 | 0.125561 |
| BABA 6h | 6118.84 | 1203.66 | 1.47652 | 0.229251 |
| Chitin 6h | 4176.14 | 298.752 | 1.09242 | 0.112601 |
| Epi 6h | 2795.73 | 161.635 | 0.693441 | 0.0403854 |
| SA 6h | 2849.77 | 676.454 | 0.758045 | 0.118175 |
| Me-JA 6h | 3252.61 | 635.926 | 0.87682 | 0.11025 |
Source Transcript PGSC0003DMT400079444 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G25490.1 | +2 | 0.0 | 622 | 335/640 (52%) | EIN3-binding F box protein 1 | chr2:10848018-10850275 REVERSE LENGTH=628 |
