Probe CUST_34987_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34987_PI426222305 | JHI_St_60k_v1 | DMT400053731 | TCTTCTCATGACACAATTCCCGAAAATCTTGAATTCCAACGTCTCGAAAATGATCCTCAG |
All Microarray Probes Designed to Gene DMG400020844
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34987_PI426222305 | JHI_St_60k_v1 | DMT400053731 | TCTTCTCATGACACAATTCCCGAAAATCTTGAATTCCAACGTCTCGAAAATGATCCTCAG |
Microarray Signals from CUST_34987_PI426222305
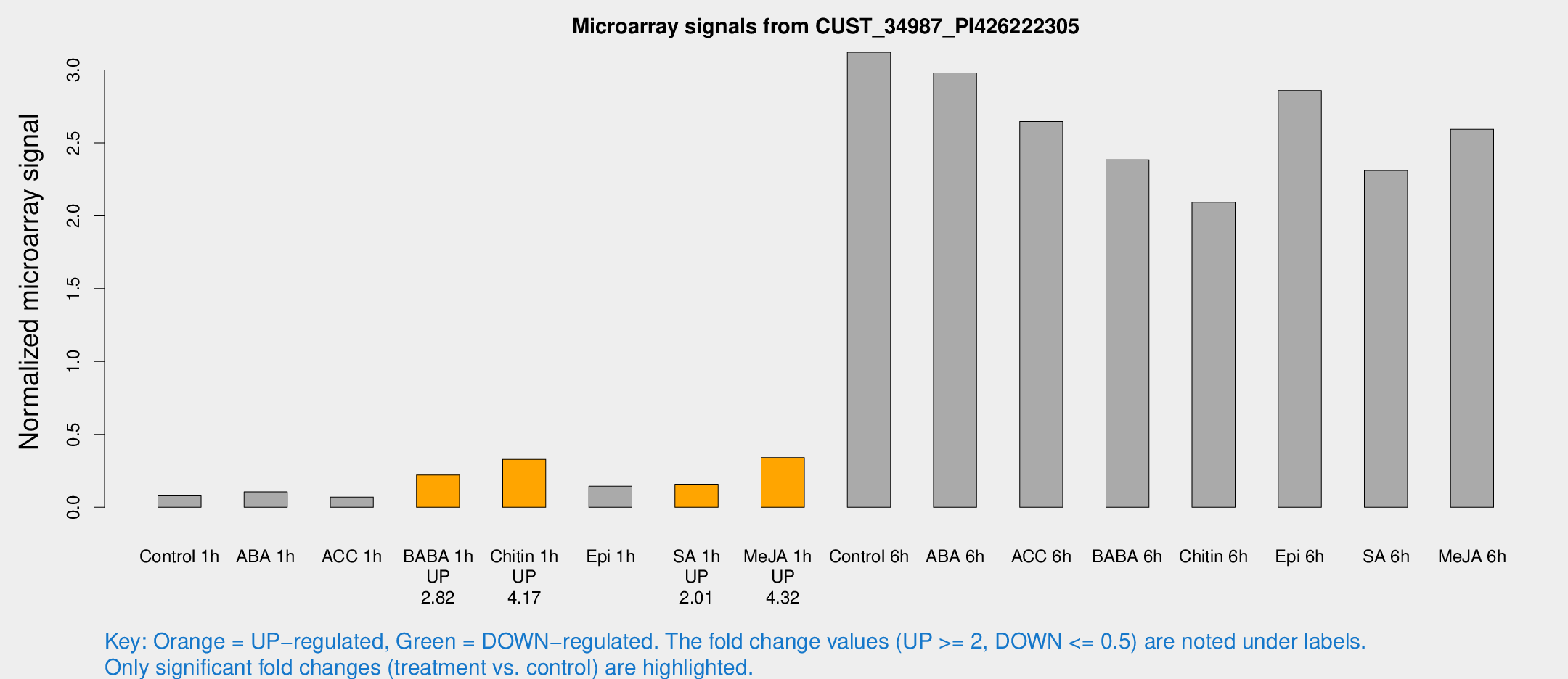
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 216.52 | 42.2325 | 0.0788809 | 0.0122423 |
| ABA 1h | 265.186 | 68.46 | 0.10558 | 0.021669 |
| ACC 1h | 203.043 | 46.8436 | 0.070193 | 0.0135209 |
| BABA 1h | 587.997 | 104.749 | 0.222114 | 0.0210383 |
| Chitin 1h | 790.452 | 74.7096 | 0.329082 | 0.0336362 |
| Epi 1h | 344.625 | 67.4759 | 0.144221 | 0.0279464 |
| SA 1h | 446.036 | 83.0669 | 0.158389 | 0.0270511 |
| Me-JA 1h | 749.373 | 99.9048 | 0.341081 | 0.0267844 |
| Control 6h | 9650.87 | 3098.51 | 3.12141 | 1.12731 |
| ABA 6h | 8488.85 | 939.463 | 2.97951 | 0.172026 |
| ACC 6h | 8472.45 | 2016.38 | 2.64647 | 0.240735 |
| BABA 6h | 7085.14 | 410.345 | 2.38413 | 0.137653 |
| Chitin 6h | 5900.65 | 340.939 | 2.0929 | 0.120841 |
| Epi 6h | 8892.64 | 1829.97 | 2.85938 | 0.700937 |
| SA 6h | 6266.22 | 1160.05 | 2.30996 | 0.238271 |
| Me-JA 6h | 7011.21 | 1087.47 | 2.5926 | 0.202554 |
Source Transcript PGSC0003DMT400053731 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G37300.1 | +3 | 4e-24 | 98 | 72/183 (39%) | maternal effect embryo arrest 59 | chr4:17554805-17555498 FORWARD LENGTH=173 |
