Probe CUST_34810_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34810_PI426222305 | JHI_St_60k_v1 | DMT400095174 | ATATTTGAGCGCGAGACACCTCTGGCAGATAGTGATTCTGTCAGAGCAAGAACATTGGAT |
All Microarray Probes Designed to Gene DMG400044745
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34810_PI426222305 | JHI_St_60k_v1 | DMT400095174 | ATATTTGAGCGCGAGACACCTCTGGCAGATAGTGATTCTGTCAGAGCAAGAACATTGGAT |
Microarray Signals from CUST_34810_PI426222305
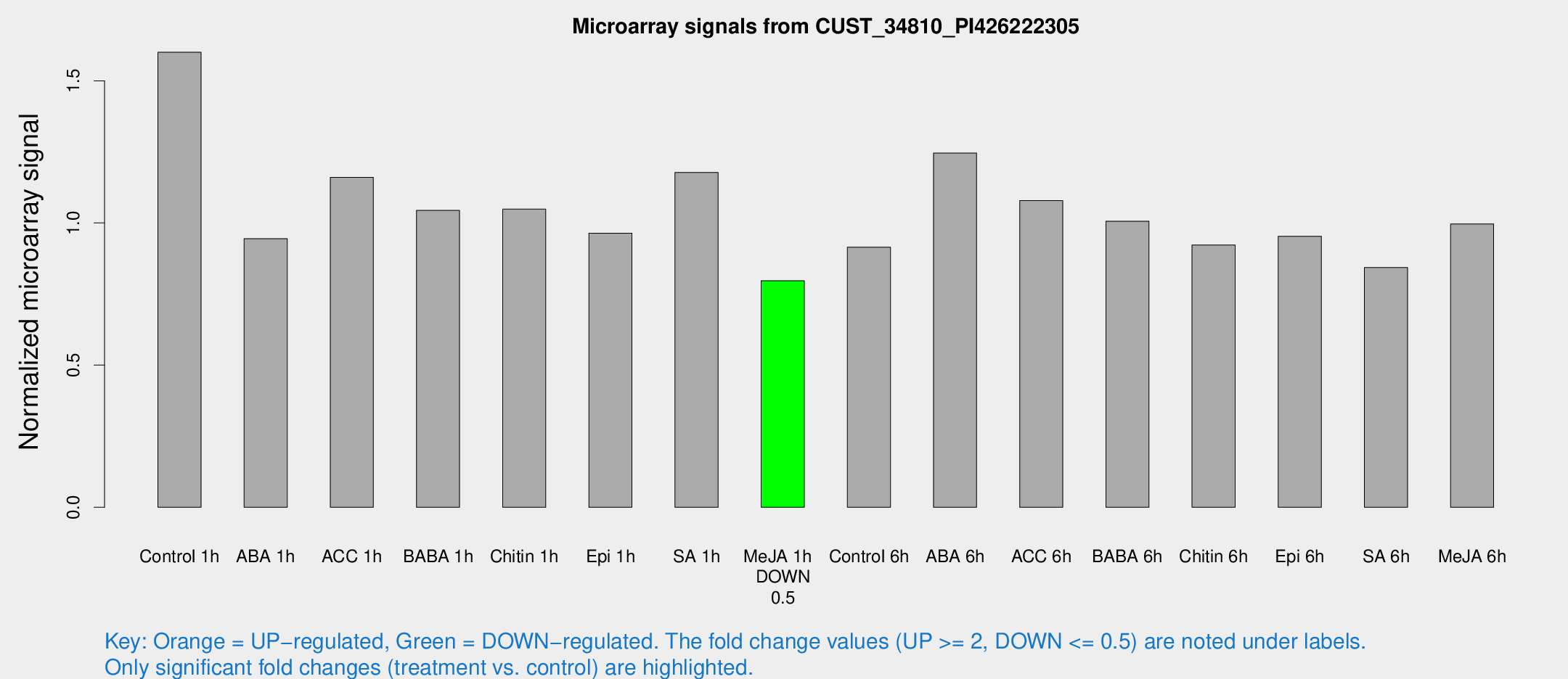
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2048.92 | 185.821 | 1.60051 | 0.0924328 |
| ABA 1h | 1061.88 | 61.4451 | 0.944801 | 0.0546018 |
| ACC 1h | 1621.26 | 377.931 | 1.16063 | 0.238063 |
| BABA 1h | 1316.92 | 228.935 | 1.04428 | 0.103267 |
| Chitin 1h | 1208.94 | 123.711 | 1.04851 | 0.101926 |
| Epi 1h | 1059.33 | 61.2636 | 0.963905 | 0.0557108 |
| SA 1h | 1563.26 | 208.875 | 1.17785 | 0.136559 |
| Me-JA 1h | 832.256 | 74.9587 | 0.796772 | 0.0460858 |
| Control 6h | 1213.97 | 228.96 | 0.91498 | 0.113321 |
| ABA 6h | 1706.43 | 202.385 | 1.24583 | 0.0944323 |
| ACC 6h | 1589.81 | 172.126 | 1.07884 | 0.113312 |
| BABA 6h | 1451.2 | 171.985 | 1.00628 | 0.0859232 |
| Chitin 6h | 1248.22 | 72.2473 | 0.922714 | 0.061971 |
| Epi 6h | 1388.89 | 183.593 | 0.953377 | 0.180445 |
| SA 6h | 1073 | 119.625 | 0.843324 | 0.0487572 |
| Me-JA 6h | 1284.34 | 169.799 | 0.996667 | 0.0740832 |
Source Transcript PGSC0003DMT400095174 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G48010.1 | +1 | 5e-115 | 347 | 179/253 (71%) | receptor-like kinase in in flowers 3 | chr2:19641465-19643318 FORWARD LENGTH=617 |
