Probe CUST_34466_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34466_PI426222305 | JHI_St_60k_v1 | DMT400071863 | CTATGGATGGGTTAGAATGGCTCAAGTCTAGTAAGGATGAATGGGTTTGTAATTATGCTG |
All Microarray Probes Designed to Gene DMG400027960
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34466_PI426222305 | JHI_St_60k_v1 | DMT400071863 | CTATGGATGGGTTAGAATGGCTCAAGTCTAGTAAGGATGAATGGGTTTGTAATTATGCTG |
Microarray Signals from CUST_34466_PI426222305
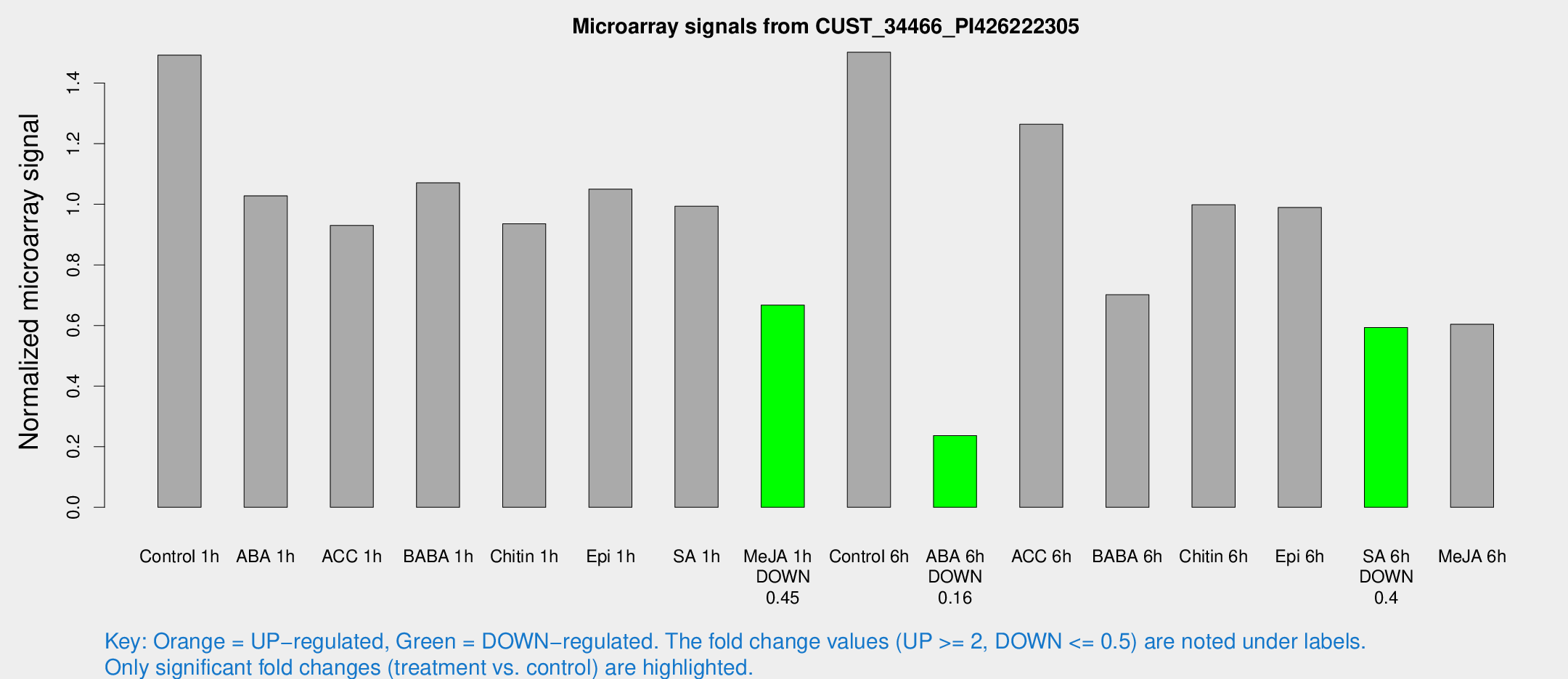
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 210.832 | 17.1444 | 1.492 | 0.0890917 |
| ABA 1h | 129.037 | 13.107 | 1.02796 | 0.0643999 |
| ACC 1h | 141.032 | 29.2102 | 0.930253 | 0.144908 |
| BABA 1h | 159.58 | 45.5316 | 1.07062 | 0.250185 |
| Chitin 1h | 120.47 | 16.1748 | 0.935945 | 0.0623686 |
| Epi 1h | 129.473 | 16.144 | 1.04999 | 0.141049 |
| SA 1h | 151.254 | 33.343 | 0.993683 | 0.17948 |
| Me-JA 1h | 79.7486 | 17.0028 | 0.667233 | 0.0827552 |
| Control 6h | 232.919 | 69.301 | 1.50182 | 0.386045 |
| ABA 6h | 35.3683 | 3.92769 | 0.236803 | 0.0262664 |
| ACC 6h | 217.454 | 57.2462 | 1.26412 | 0.152277 |
| BABA 6h | 119.988 | 35.3285 | 0.701411 | 0.198773 |
| Chitin 6h | 151.009 | 16.0251 | 0.998471 | 0.104717 |
| Epi 6h | 163.848 | 31.8539 | 0.989288 | 0.297657 |
| SA 6h | 86.8716 | 17.9954 | 0.593356 | 0.0724229 |
| Me-JA 6h | 96.543 | 32.7632 | 0.603964 | 0.190184 |
Source Transcript PGSC0003DMT400071863 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G62180.1 | +1 | 2e-91 | 280 | 135/313 (43%) | carboxyesterase 20 | chr5:24978866-24979849 REVERSE LENGTH=327 |
