Probe CUST_34411_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34411_PI426222305 | JHI_St_60k_v1 | DMT400035638 | CATTTCCCATGGACACAGGTACGCACGCTACGCCTTCTTATTAGTAATAAACTGTTCCTT |
All Microarray Probes Designed to Gene DMG401013710
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34411_PI426222305 | JHI_St_60k_v1 | DMT400035638 | CATTTCCCATGGACACAGGTACGCACGCTACGCCTTCTTATTAGTAATAAACTGTTCCTT |
Microarray Signals from CUST_34411_PI426222305
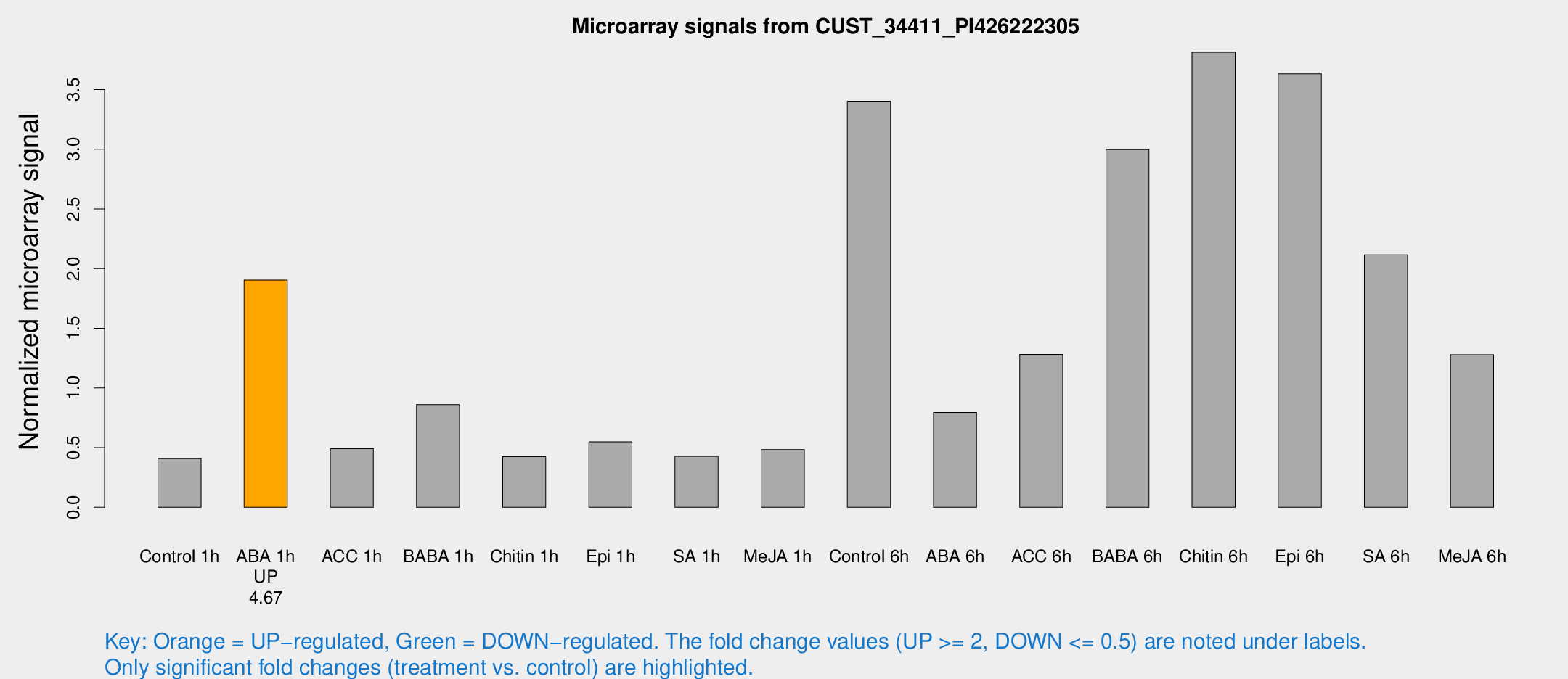
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.6869 | 3.04819 | 0.40766 | 0.202292 |
| ABA 1h | 26.7804 | 3.31337 | 1.90435 | 0.24156 |
| ACC 1h | 8.81608 | 3.32635 | 0.491673 | 0.231257 |
| BABA 1h | 14.3829 | 3.82041 | 0.860933 | 0.257132 |
| Chitin 1h | 6.0638 | 3.07543 | 0.425046 | 0.219921 |
| Epi 1h | 8.09247 | 3.08953 | 0.548478 | 0.249577 |
| SA 1h | 7.46884 | 3.06958 | 0.427644 | 0.203174 |
| Me-JA 1h | 6.32183 | 3.21814 | 0.48402 | 0.253394 |
| Control 6h | 61.0714 | 19.3057 | 3.40297 | 1.06678 |
| ABA 6h | 14.3386 | 3.69108 | 0.795561 | 0.219534 |
| ACC 6h | 30.1218 | 15.832 | 1.28171 | 0.52225 |
| BABA 6h | 54.696 | 9.66772 | 2.99652 | 0.482503 |
| Chitin 6h | 65.9302 | 11.798 | 3.81311 | 0.651533 |
| Epi 6h | 69.849 | 19.1393 | 3.63195 | 1.2768 |
| SA 6h | 39.0931 | 15.5778 | 2.11603 | 1.05091 |
| Me-JA 6h | 28.5723 | 12.3755 | 1.27864 | 1.04963 |
Source Transcript PGSC0003DMT400035638 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18910.1 | +1 | 1e-19 | 83 | 38/80 (48%) | NOD26-like intrinsic protein 1;2 | chr4:10366211-10368179 FORWARD LENGTH=294 |
