Probe CUST_34153_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34153_PI426222305 | JHI_St_60k_v1 | DMT400033082 | CAAGGCCTTGTTCGGCTGCATATTTGTATGTAATTGTACTTTTTCATGAATTCGTTGTAT |
All Microarray Probes Designed to Gene DMG400012704
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34153_PI426222305 | JHI_St_60k_v1 | DMT400033082 | CAAGGCCTTGTTCGGCTGCATATTTGTATGTAATTGTACTTTTTCATGAATTCGTTGTAT |
Microarray Signals from CUST_34153_PI426222305
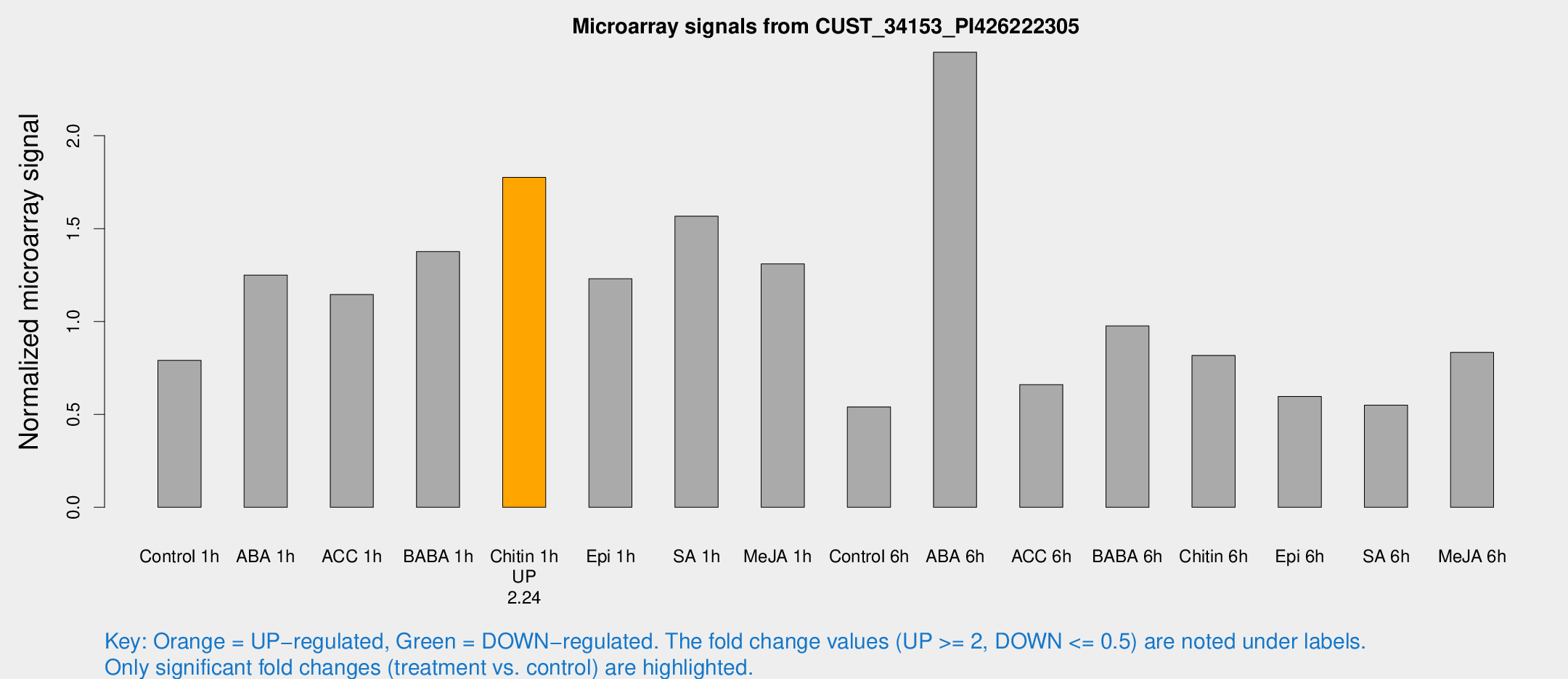
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2129.74 | 181.238 | 0.791189 | 0.0456919 |
| ABA 1h | 3010.74 | 393.014 | 1.25015 | 0.0944926 |
| ACC 1h | 3155.68 | 204.736 | 1.14491 | 0.162201 |
| BABA 1h | 3881.58 | 1045.72 | 1.37618 | 0.305087 |
| Chitin 1h | 4356.52 | 621.131 | 1.77589 | 0.225259 |
| Epi 1h | 2849.11 | 164.706 | 1.23019 | 0.0725422 |
| SA 1h | 4535.63 | 1091.63 | 1.5669 | 0.27154 |
| Me-JA 1h | 2902.36 | 347.409 | 1.31016 | 0.125485 |
| Control 6h | 1572.56 | 394.995 | 0.540353 | 0.118776 |
| ABA 6h | 7379.19 | 1832.05 | 2.44936 | 0.460408 |
| ACC 6h | 2108.18 | 429.785 | 0.660513 | 0.0467371 |
| BABA 6h | 2934.69 | 194.913 | 0.976334 | 0.0563796 |
| Chitin 6h | 2328.79 | 134.639 | 0.817596 | 0.052894 |
| Epi 6h | 1805.55 | 125.879 | 0.596253 | 0.0439959 |
| SA 6h | 1704.99 | 557.439 | 0.549585 | 0.189089 |
| Me-JA 6h | 2353.62 | 531.341 | 0.834011 | 0.165672 |
Source Transcript PGSC0003DMT400033082 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G36430.1 | +2 | 3e-25 | 109 | 110/421 (26%) | Plant protein of unknown function (DUF247) | chr2:15290211-15291557 FORWARD LENGTH=448 |
