Probe CUST_33971_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33971_PI426222305 | JHI_St_60k_v1 | DMT400062174 | TTGACAGTAATTGGGTACCAACACCAAGCTGATGTAGATGCTGGAGGTGATGTATGTGGT |
All Microarray Probes Designed to Gene DMG400024197
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33971_PI426222305 | JHI_St_60k_v1 | DMT400062174 | TTGACAGTAATTGGGTACCAACACCAAGCTGATGTAGATGCTGGAGGTGATGTATGTGGT |
| CUST_33993_PI426222305 | JHI_St_60k_v1 | DMT400062173 | TCCCACGTACCCGTAAGTATATATATTCAGAGTTTAAATTCTATGGAGTGACGTGACAAT |
Microarray Signals from CUST_33971_PI426222305
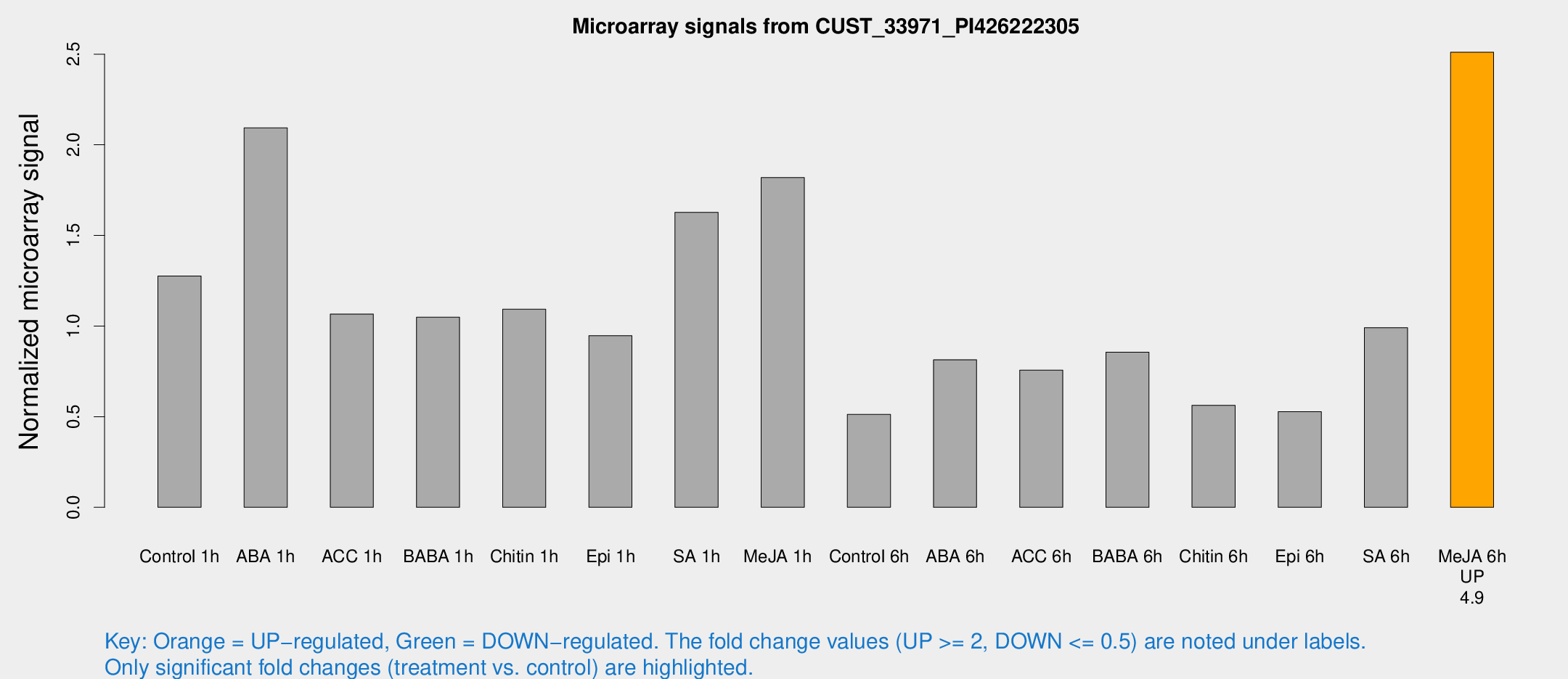
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 741.119 | 99.8412 | 1.27576 | 0.102727 |
| ABA 1h | 1072.48 | 140.981 | 2.0932 | 0.124928 |
| ACC 1h | 643.119 | 106.26 | 1.06601 | 0.115949 |
| BABA 1h | 609.91 | 136.778 | 1.04888 | 0.158116 |
| Chitin 1h | 568.332 | 68.9053 | 1.09283 | 0.112604 |
| Epi 1h | 470.456 | 39.5097 | 0.947188 | 0.0765323 |
| SA 1h | 967.284 | 115.407 | 1.62745 | 0.115699 |
| Me-JA 1h | 856.819 | 99.5036 | 1.81912 | 0.105288 |
| Control 6h | 308.812 | 66.1506 | 0.512186 | 0.0813545 |
| ABA 6h | 493.847 | 28.8086 | 0.813926 | 0.053514 |
| ACC 6h | 513.299 | 91.6815 | 0.75687 | 0.137361 |
| BABA 6h | 548.936 | 42.6781 | 0.855316 | 0.0836443 |
| Chitin 6h | 342.498 | 20.7374 | 0.5628 | 0.0403124 |
| Epi 6h | 348.72 | 55.1433 | 0.527641 | 0.0612417 |
| SA 6h | 606.74 | 162.817 | 0.990987 | 0.195198 |
| Me-JA 6h | 1429.43 | 83.9069 | 2.51073 | 0.269438 |
Source Transcript PGSC0003DMT400062174 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37170.1 | +1 | 1e-160 | 456 | 236/286 (83%) | plasma membrane intrinsic protein 2 | chr2:15613624-15614791 REVERSE LENGTH=285 |
