Probe CUST_33225_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33225_PI426222305 | JHI_St_60k_v1 | DMT400067617 | GCTTATTGTGATAAAGTTGTGGGTTATGGACCTGAAGCTGTTAAAGGACAATTGATTAAG |
All Microarray Probes Designed to Gene DMG400026290
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33225_PI426222305 | JHI_St_60k_v1 | DMT400067617 | GCTTATTGTGATAAAGTTGTGGGTTATGGACCTGAAGCTGTTAAAGGACAATTGATTAAG |
Microarray Signals from CUST_33225_PI426222305
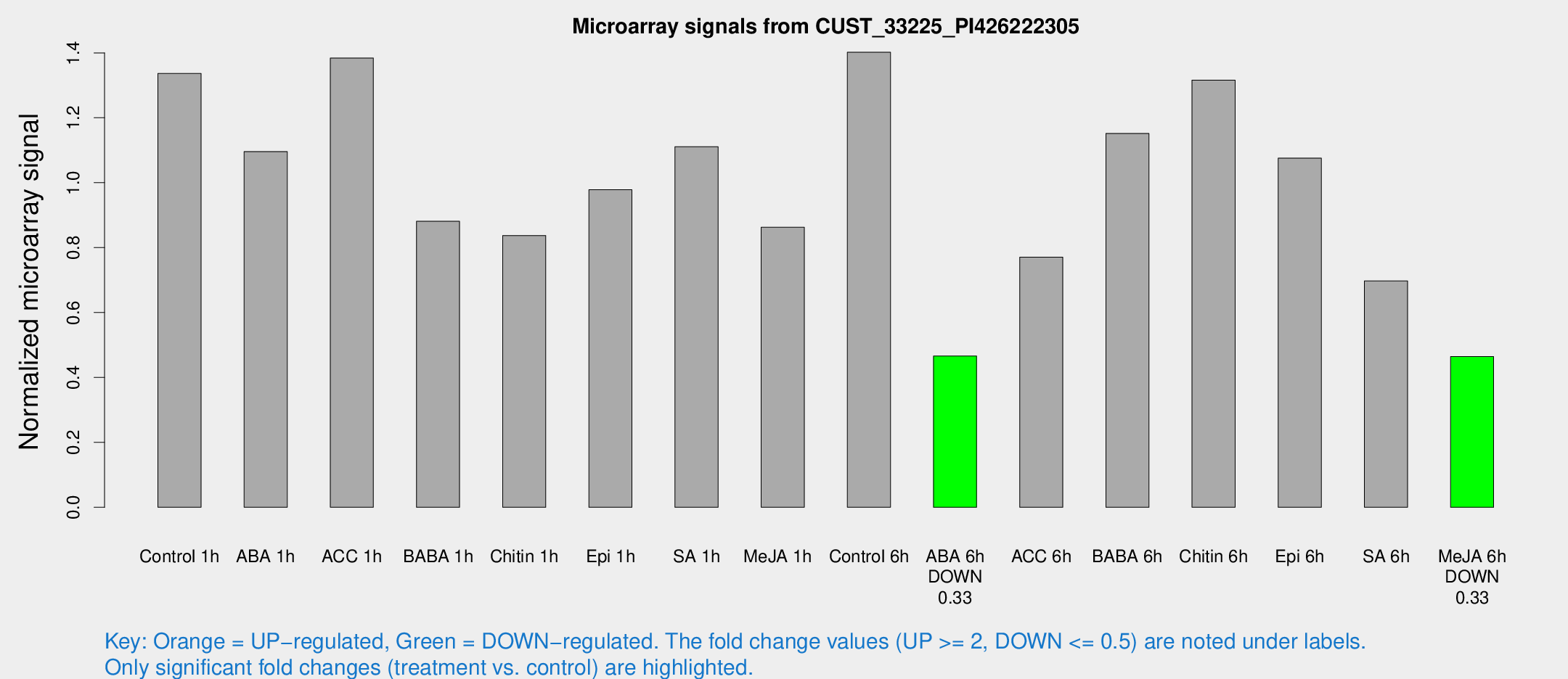
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1812.7 | 287.02 | 1.33648 | 0.126662 |
| ABA 1h | 1297.78 | 139.882 | 1.09602 | 0.126524 |
| ACC 1h | 1899.82 | 201.643 | 1.38388 | 0.07998 |
| BABA 1h | 1160.57 | 195.196 | 0.881124 | 0.0722994 |
| Chitin 1h | 1024.15 | 179.945 | 0.837334 | 0.158264 |
| Epi 1h | 1123.09 | 65.1529 | 0.978291 | 0.0565654 |
| SA 1h | 1509.84 | 87.3025 | 1.11098 | 0.0641965 |
| Me-JA 1h | 932.077 | 53.9951 | 0.862699 | 0.0499257 |
| Control 6h | 1945 | 379.762 | 1.40197 | 0.181012 |
| ABA 6h | 655.453 | 38.0596 | 0.465984 | 0.027039 |
| ACC 6h | 1261.51 | 336.635 | 0.770431 | 0.132168 |
| BABA 6h | 1761.75 | 330.25 | 1.15176 | 0.218087 |
| Chitin 6h | 1902.94 | 326.306 | 1.31568 | 0.183629 |
| Epi 6h | 1632.36 | 207.829 | 1.07573 | 0.231278 |
| SA 6h | 1094.04 | 390.59 | 0.697189 | 0.26748 |
| Me-JA 6h | 670.276 | 190.862 | 0.4642 | 0.110272 |
Source Transcript PGSC0003DMT400067617 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G45680.1 | +3 | 8e-60 | 200 | 106/123 (86%) | FK506-binding protein 13 | chr5:18530894-18532128 FORWARD LENGTH=208 |
