Probe CUST_33038_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33038_PI426222305 | JHI_St_60k_v1 | DMT400016056 | ACGGTTGATCAACTTTGGAGTGATTACAAGGCTAAATTTGGGGCATAACTTAAGAAATCC |
All Microarray Probes Designed to Gene DMG400006276
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33038_PI426222305 | JHI_St_60k_v1 | DMT400016056 | ACGGTTGATCAACTTTGGAGTGATTACAAGGCTAAATTTGGGGCATAACTTAAGAAATCC |
Microarray Signals from CUST_33038_PI426222305
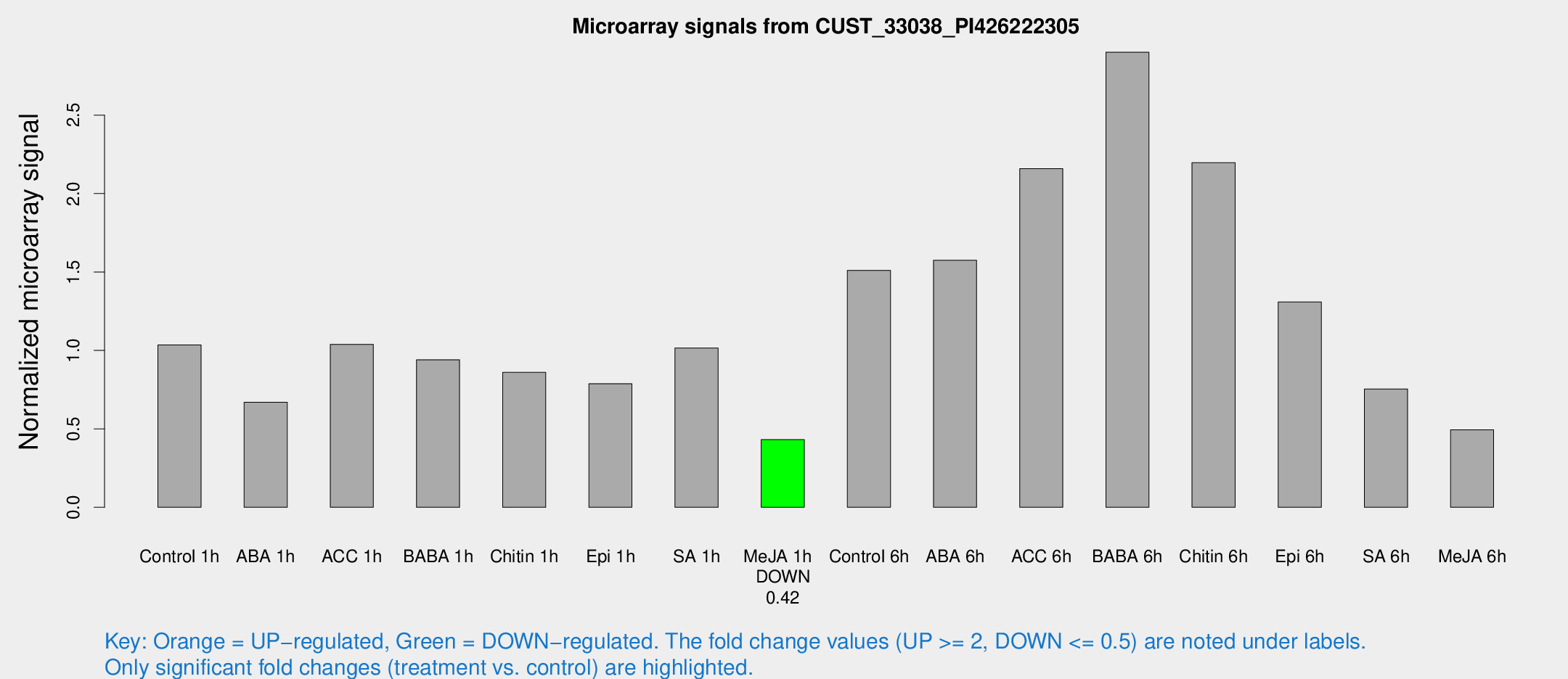
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5009.35 | 843.499 | 1.03498 | 0.103945 |
| ABA 1h | 2999.91 | 852.926 | 0.669606 | 0.137578 |
| ACC 1h | 5861.04 | 2037.86 | 1.0392 | 0.384653 |
| BABA 1h | 4364.98 | 632.223 | 0.940539 | 0.0563151 |
| Chitin 1h | 3685.86 | 398.274 | 0.860476 | 0.0807595 |
| Epi 1h | 3248.18 | 352.391 | 0.788183 | 0.067043 |
| SA 1h | 4918.73 | 284.51 | 1.01542 | 0.10337 |
| Me-JA 1h | 1678.57 | 191.516 | 0.431452 | 0.0249234 |
| Control 6h | 8553.35 | 3310.57 | 1.51059 | 0.593619 |
| ABA 6h | 8221.09 | 1767.89 | 1.57466 | 0.250433 |
| ACC 6h | 12003.9 | 2033.24 | 2.15931 | 0.230493 |
| BABA 6h | 15503.7 | 1824.46 | 2.90125 | 0.231062 |
| Chitin 6h | 11157.8 | 1265.52 | 2.19695 | 0.31375 |
| Epi 6h | 7532.85 | 1938.98 | 1.30875 | 0.540032 |
| SA 6h | 3577.08 | 493.061 | 0.754577 | 0.0812847 |
| Me-JA 6h | 2929.8 | 1420.58 | 0.494599 | 0.222668 |
Source Transcript PGSC0003DMT400016056 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G15220.1 | +2 | 5e-104 | 306 | 143/224 (64%) | Plant basic secretory protein (BSP) family protein | chr2:6608689-6609366 FORWARD LENGTH=225 |
