Probe CUST_32779_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32779_PI426222305 | JHI_St_60k_v1 | DMT400047315 | CAATCTGAAGAATCATTGAGAAAAGTTATGTATTTGAGCTGTTGGGGTCCAAATTGATTG |
All Microarray Probes Designed to Gene DMG400018377
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32779_PI426222305 | JHI_St_60k_v1 | DMT400047315 | CAATCTGAAGAATCATTGAGAAAAGTTATGTATTTGAGCTGTTGGGGTCCAAATTGATTG |
Microarray Signals from CUST_32779_PI426222305
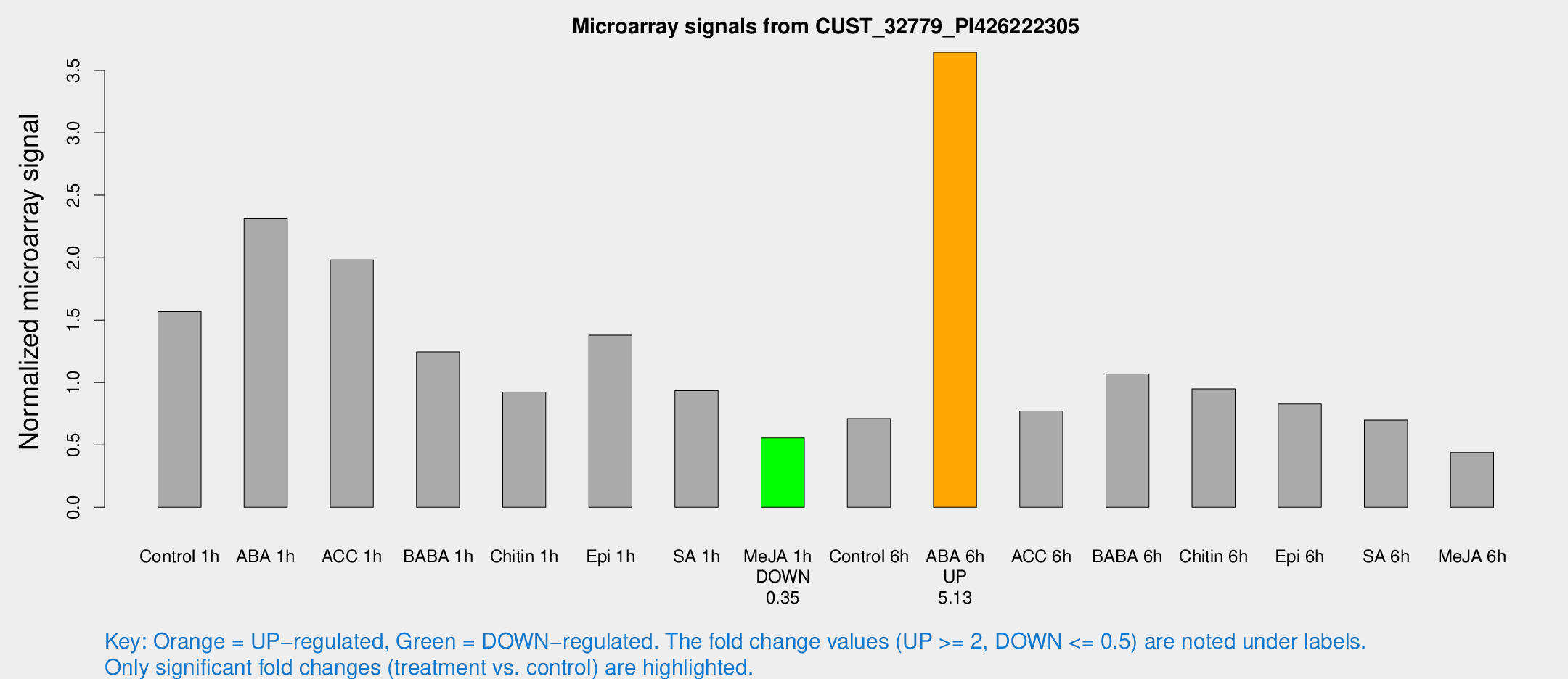
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 122.672 | 27.2895 | 1.56741 | 0.284242 |
| ABA 1h | 160.911 | 34.5578 | 2.31244 | 0.473988 |
| ACC 1h | 162.681 | 41.5753 | 1.98222 | 0.418184 |
| BABA 1h | 90.1837 | 7.57199 | 1.24488 | 0.143372 |
| Chitin 1h | 63.4439 | 10.2091 | 0.922652 | 0.0775773 |
| Epi 1h | 92.2773 | 17.2177 | 1.37976 | 0.225875 |
| SA 1h | 75.5944 | 15.9656 | 0.934812 | 0.232833 |
| Me-JA 1h | 33.8511 | 4.01364 | 0.555021 | 0.0655748 |
| Control 6h | 59.3564 | 16.7049 | 0.710404 | 0.197288 |
| ABA 6h | 291.987 | 28.8148 | 3.64538 | 0.407416 |
| ACC 6h | 67.4848 | 9.85594 | 0.771903 | 0.0654337 |
| BABA 6h | 91.2437 | 13.9702 | 1.06844 | 0.128496 |
| Chitin 6h | 75.5452 | 5.8854 | 0.948786 | 0.0739494 |
| Epi 6h | 70.06 | 5.76346 | 0.828305 | 0.121702 |
| SA 6h | 54.3821 | 11.8199 | 0.698626 | 0.213261 |
| Me-JA 6h | 34.8344 | 7.95182 | 0.439399 | 0.0834246 |
Source Transcript PGSC0003DMT400047315 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10265.1 | +1 | 9e-34 | 116 | 54/83 (65%) | Wound-responsive family protein | chr4:6373226-6373477 REVERSE LENGTH=83 |
