Probe CUST_32761_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32761_PI426222305 | JHI_St_60k_v1 | DMT400047341 | CTCTTAGGAAAGTTATCTACTTGACCTGTTGGGGTCCATATTGAATTCCAACTGTTAAAT |
All Microarray Probes Designed to Gene DMG400018398
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32761_PI426222305 | JHI_St_60k_v1 | DMT400047341 | CTCTTAGGAAAGTTATCTACTTGACCTGTTGGGGTCCATATTGAATTCCAACTGTTAAAT |
Microarray Signals from CUST_32761_PI426222305
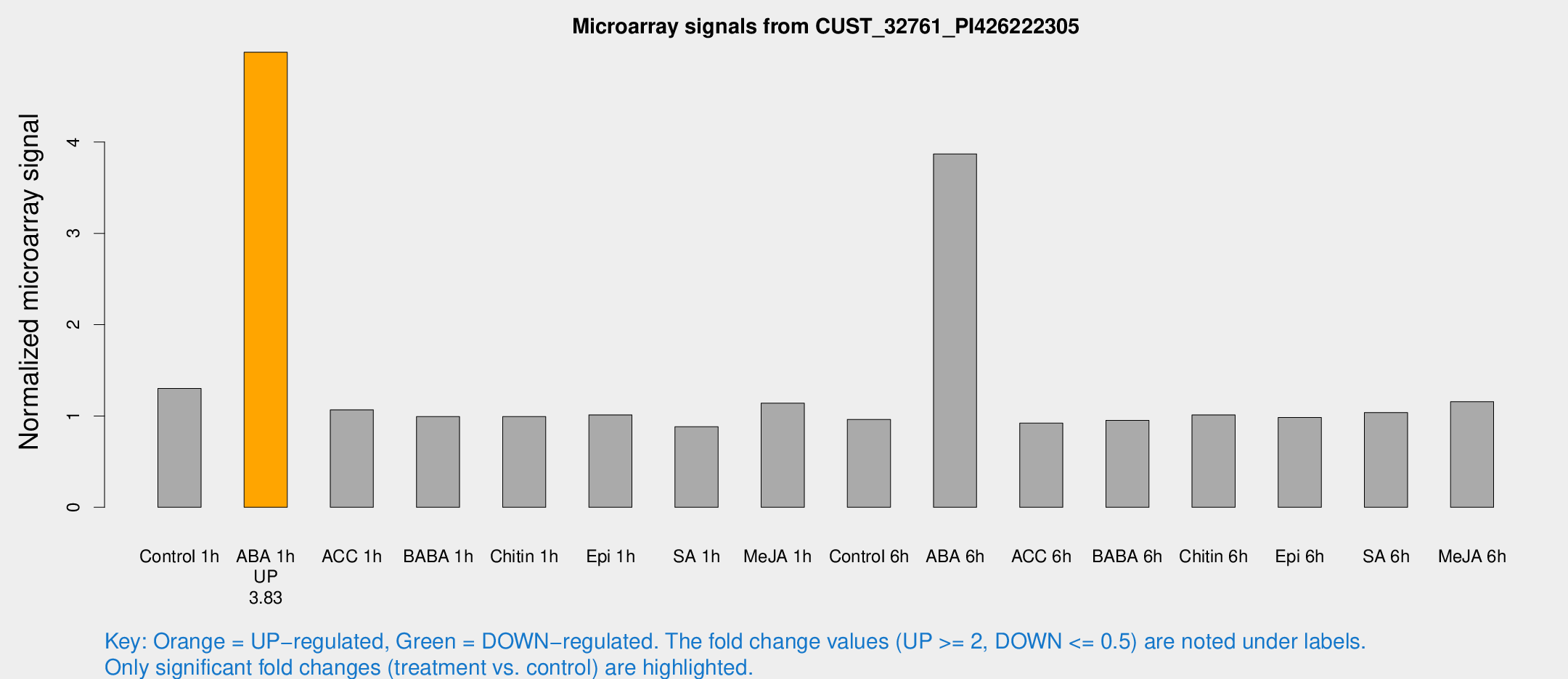
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.36314 | 3.80505 | 1.30224 | 0.565289 |
| ABA 1h | 31.7424 | 4.82085 | 4.98393 | 0.722839 |
| ACC 1h | 7.87666 | 4.66409 | 1.06757 | 0.619159 |
| BABA 1h | 6.77313 | 3.94235 | 0.993064 | 0.57504 |
| Chitin 1h | 6.33641 | 3.69625 | 0.993594 | 0.575412 |
| Epi 1h | 6.21305 | 3.62311 | 1.01237 | 0.586518 |
| SA 1h | 6.39356 | 3.71539 | 0.882055 | 0.511471 |
| Me-JA 1h | 6.60746 | 3.86266 | 1.14033 | 0.660927 |
| Control 6h | 6.80943 | 3.95296 | 0.962798 | 0.557687 |
| ABA 6h | 32.5906 | 9.77865 | 3.87008 | 1.86694 |
| ACC 6h | 7.59549 | 4.49175 | 0.921579 | 0.533659 |
| BABA 6h | 7.53931 | 4.3862 | 0.952431 | 0.551572 |
| Chitin 6h | 7.6065 | 4.416 | 1.01231 | 0.586151 |
| Epi 6h | 8.00183 | 4.75499 | 0.983665 | 0.569788 |
| SA 6h | 7.23076 | 4.18927 | 1.03778 | 0.600959 |
| Me-JA 6h | 8.35748 | 3.94483 | 1.15698 | 0.582035 |
Source Transcript PGSC0003DMT400047341 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10270.1 | +2 | 2e-30 | 108 | 53/89 (60%) | Wound-responsive family protein | chr4:6374805-6375077 FORWARD LENGTH=90 |
