Probe CUST_32431_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32431_PI426222305 | JHI_St_60k_v1 | DMT400079099 | GAAGAAAATCTTTTTGGTAGCGCCACCTCAAATAGGCCTGATGTGAGCTCTGAACAGGAA |
All Microarray Probes Designed to Gene DMG400030784
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32431_PI426222305 | JHI_St_60k_v1 | DMT400079099 | GAAGAAAATCTTTTTGGTAGCGCCACCTCAAATAGGCCTGATGTGAGCTCTGAACAGGAA |
Microarray Signals from CUST_32431_PI426222305
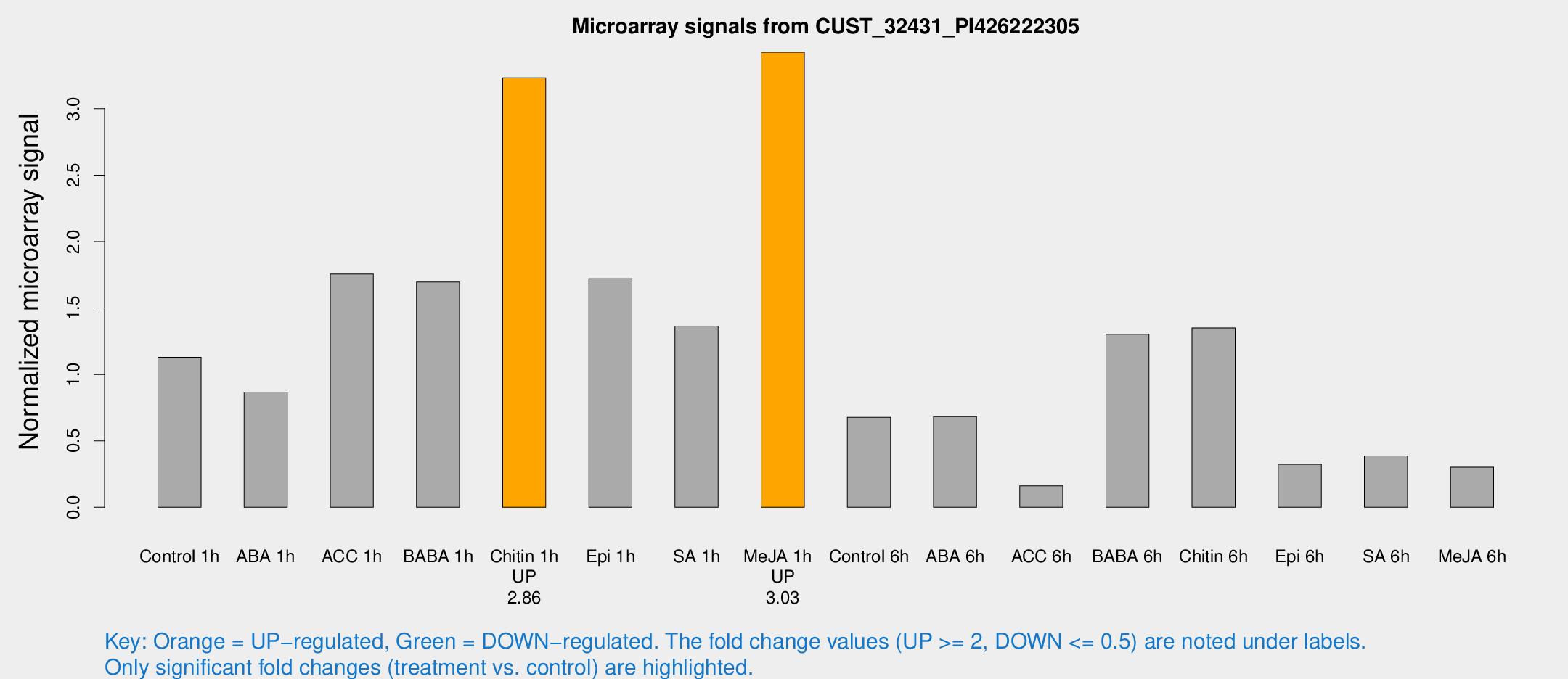
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 112.193 | 24.3663 | 1.12899 | 0.211568 |
| ABA 1h | 81.5112 | 30.0423 | 0.867055 | 0.257275 |
| ACC 1h | 189.058 | 54.4467 | 1.75526 | 0.803033 |
| BABA 1h | 155.736 | 18.7094 | 1.69517 | 0.146284 |
| Chitin 1h | 280.901 | 47.6744 | 3.23184 | 0.515687 |
| Epi 1h | 142.738 | 19.5874 | 1.72068 | 0.21292 |
| SA 1h | 137.275 | 26.3372 | 1.3633 | 0.289699 |
| Me-JA 1h | 266.274 | 29.9016 | 3.42495 | 0.203877 |
| Control 6h | 75.0174 | 24.3641 | 0.677392 | 0.2576 |
| ABA 6h | 68.5446 | 5.65319 | 0.682585 | 0.0701063 |
| ACC 6h | 22.7951 | 10.92 | 0.161915 | 0.0764782 |
| BABA 6h | 146.824 | 37.9049 | 1.30292 | 0.38448 |
| Chitin 6h | 159.268 | 67.0297 | 1.35054 | 0.553164 |
| Epi 6h | 34.5995 | 4.94519 | 0.324463 | 0.046989 |
| SA 6h | 38.704 | 9.974 | 0.386813 | 0.0695791 |
| Me-JA 6h | 47.4582 | 27.3615 | 0.302677 | 0.368494 |
Source Transcript PGSC0003DMT400079099 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G15700.1 | +2 | 7e-40 | 138 | 66/103 (64%) | Thioredoxin superfamily protein | chr4:8937545-8937853 FORWARD LENGTH=102 |
