Probe CUST_32165_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32165_PI426222305 | JHI_St_60k_v1 | DMT400037628 | CAAGATTTTTGTGGGGATTTATCATCATCAAAATCTAGCTAGTGTATACACACCTTGTAC |
All Microarray Probes Designed to Gene DMG400014510
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32165_PI426222305 | JHI_St_60k_v1 | DMT400037628 | CAAGATTTTTGTGGGGATTTATCATCATCAAAATCTAGCTAGTGTATACACACCTTGTAC |
Microarray Signals from CUST_32165_PI426222305
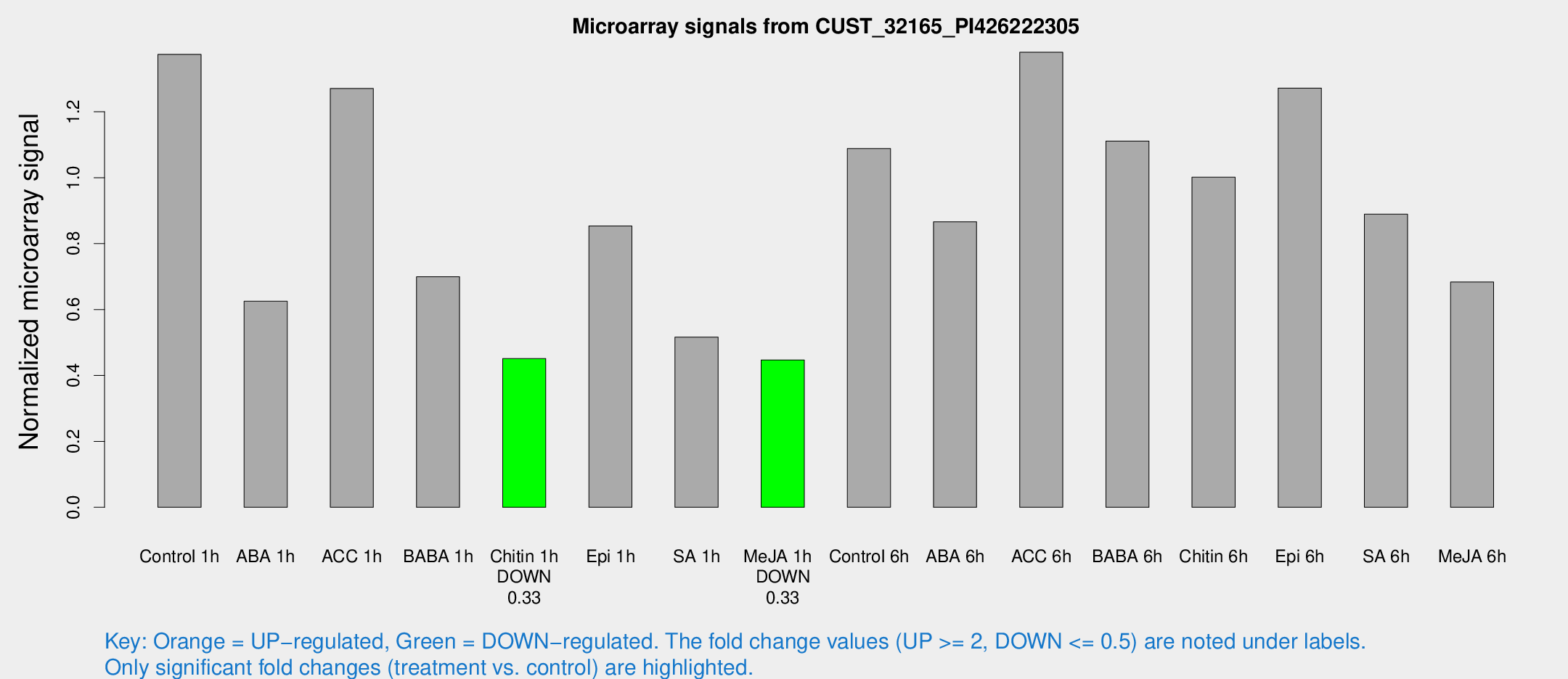
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 20.1742 | 3.20475 | 1.37421 | 0.227735 |
| ABA 1h | 8.97885 | 3.35066 | 0.625346 | 0.263785 |
| ACC 1h | 18.739 | 3.34993 | 1.27079 | 0.228862 |
| BABA 1h | 10.53 | 3.14932 | 0.699572 | 0.258891 |
| Chitin 1h | 5.82519 | 2.95548 | 0.45108 | 0.229639 |
| Epi 1h | 12.3628 | 4.97553 | 0.853472 | 0.349293 |
| SA 1h | 8.57209 | 3.16083 | 0.516434 | 0.228899 |
| Me-JA 1h | 5.15339 | 2.9911 | 0.447083 | 0.255366 |
| Control 6h | 21.0158 | 10.1375 | 1.08832 | 0.675366 |
| ABA 6h | 14.7675 | 5.09235 | 0.866202 | 0.25919 |
| ACC 6h | 22.7636 | 3.91771 | 1.38059 | 0.235148 |
| BABA 6h | 18.4327 | 3.5796 | 1.11118 | 0.234185 |
| Chitin 6h | 16.8096 | 4.62356 | 1.00144 | 0.399952 |
| Epi 6h | 20.5332 | 3.69048 | 1.27192 | 0.231861 |
| SA 6h | 15.3785 | 7.05275 | 0.889466 | 0.3557 |
| Me-JA 6h | 10.7192 | 3.09873 | 0.683585 | 0.248389 |
Source Transcript PGSC0003DMT400037628 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G25620.2 | +2 | 0.0 | 569 | 284/410 (69%) | Flavin-binding monooxygenase family protein | chr5:8935312-8938200 REVERSE LENGTH=426 |
