Probe CUST_31844_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31844_PI426222305 | JHI_St_60k_v1 | DMT400071281 | GGCCAATTCCCATTTCCATGATCCTTCTCTATTTAATGTACGTTTACGCATTTCCTTGTA |
All Microarray Probes Designed to Gene DMG400027718
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31844_PI426222305 | JHI_St_60k_v1 | DMT400071281 | GGCCAATTCCCATTTCCATGATCCTTCTCTATTTAATGTACGTTTACGCATTTCCTTGTA |
Microarray Signals from CUST_31844_PI426222305
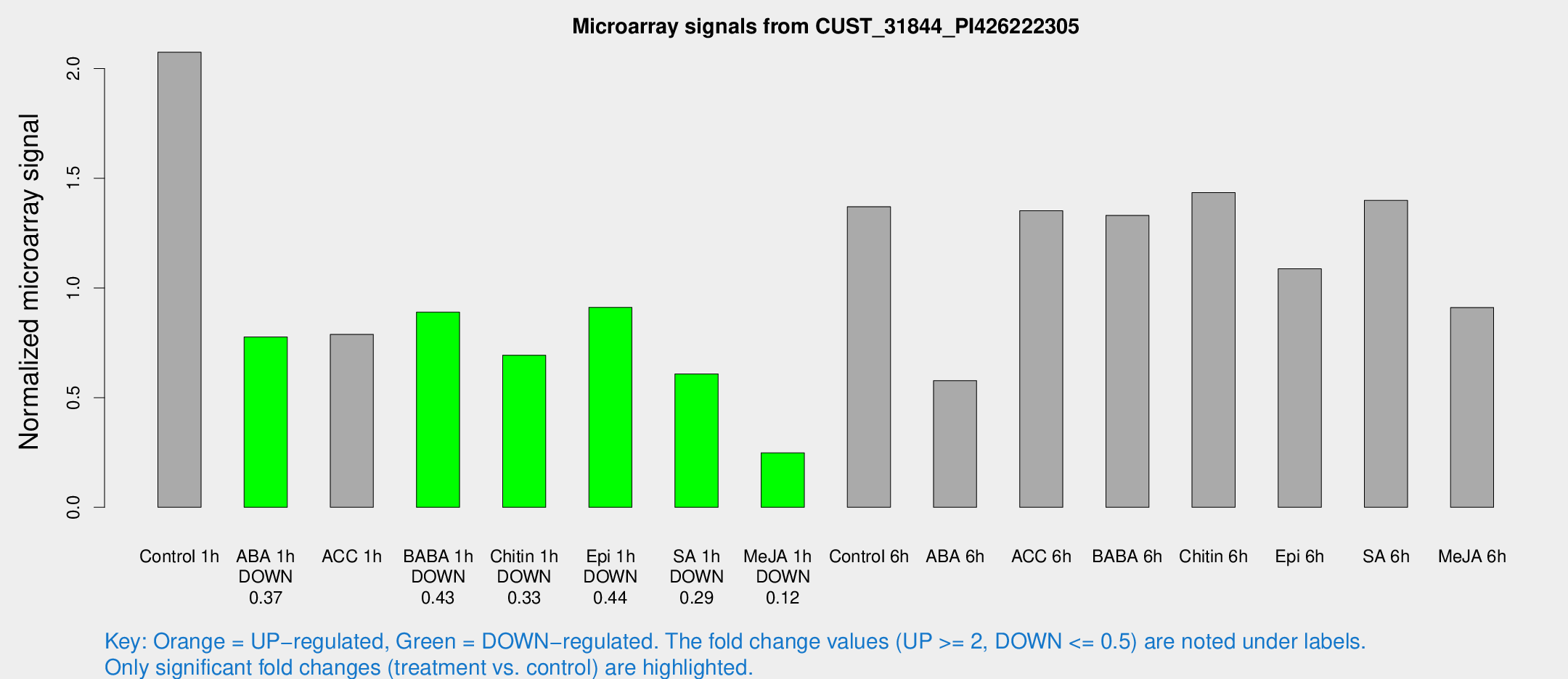
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1248.74 | 154.397 | 2.07483 | 0.122234 |
| ABA 1h | 410.585 | 37.3067 | 0.776365 | 0.0452888 |
| ACC 1h | 538.676 | 156.087 | 0.788428 | 0.242711 |
| BABA 1h | 529.191 | 97.8216 | 0.889589 | 0.093004 |
| Chitin 1h | 374.595 | 43.387 | 0.693258 | 0.0810751 |
| Epi 1h | 482.483 | 81.4181 | 0.911337 | 0.150907 |
| SA 1h | 384.198 | 69.6327 | 0.608412 | 0.0840371 |
| Me-JA 1h | 121.358 | 12.1676 | 0.248257 | 0.0268149 |
| Control 6h | 849.846 | 180.638 | 1.3705 | 0.224463 |
| ABA 6h | 368.441 | 42.6518 | 0.577442 | 0.0965146 |
| ACC 6h | 938.117 | 126.623 | 1.35205 | 0.174719 |
| BABA 6h | 901.071 | 128.941 | 1.33074 | 0.183013 |
| Chitin 6h | 914.251 | 92.6399 | 1.43477 | 0.121491 |
| Epi 6h | 754.755 | 135.906 | 1.08745 | 0.144181 |
| SA 6h | 824.328 | 58.2704 | 1.39897 | 0.0810694 |
| Me-JA 6h | 540.632 | 40.4268 | 0.910817 | 0.0529913 |
Source Transcript PGSC0003DMT400071281 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G11090.1 | +2 | 3e-53 | 175 | 85/123 (69%) | LOB domain-containing protein 21 | chr3:3475036-3475533 FORWARD LENGTH=165 |
