Probe CUST_31838_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31838_PI426222305 | JHI_St_60k_v1 | DMT400071278 | ACAGACTCACATTCTTTGAGCTAGAAGACAACTTACGGGTTCAGCTGAATCAAGTAGGTT |
All Microarray Probes Designed to Gene DMG400027717
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31847_PI426222305 | JHI_St_60k_v1 | DMT400071280 | CTAATGGGATTTCATTACTGAAGCTTCATTCAGCCATGGGATGAACCCTCATGTCTACGT |
| CUST_31838_PI426222305 | JHI_St_60k_v1 | DMT400071278 | ACAGACTCACATTCTTTGAGCTAGAAGACAACTTACGGGTTCAGCTGAATCAAGTAGGTT |
Microarray Signals from CUST_31838_PI426222305
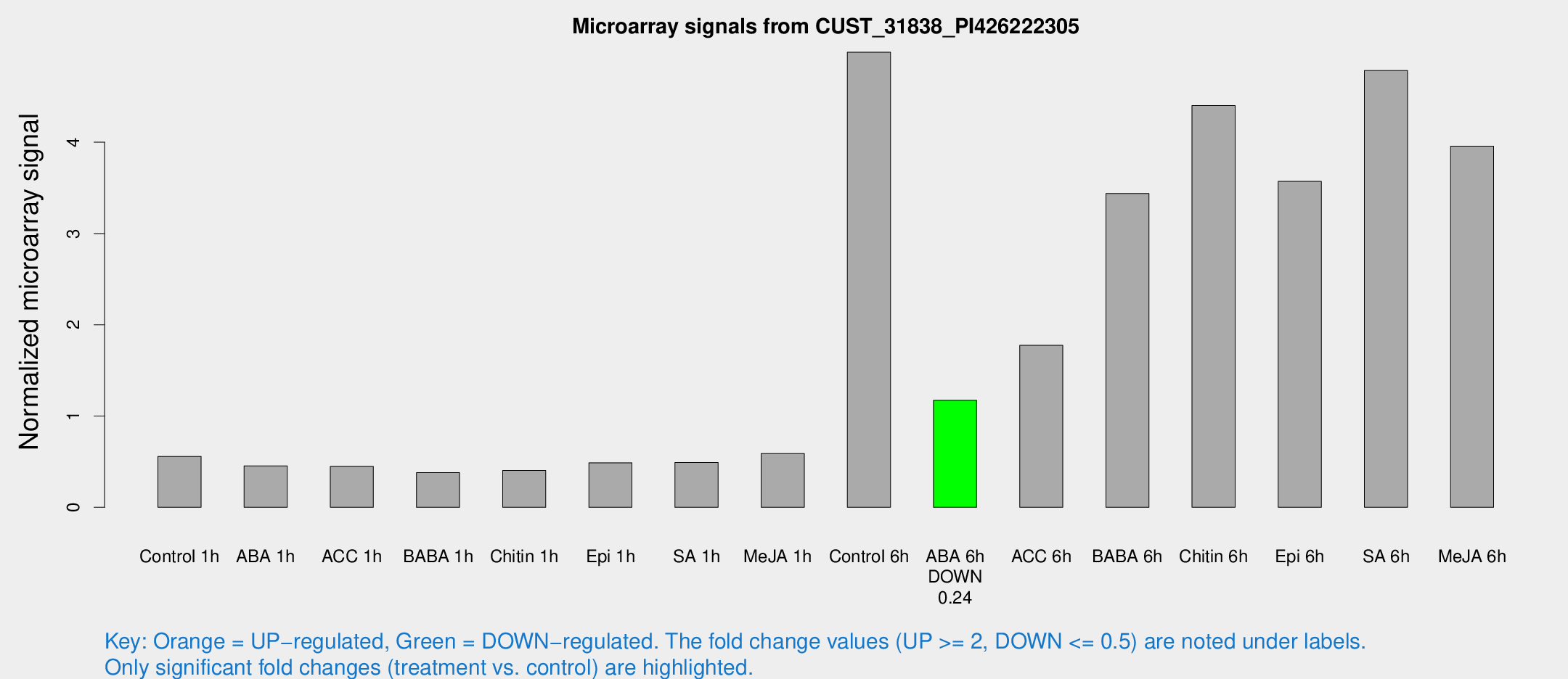
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.2677 | 16.1059 | 0.558042 | 0.184714 |
| ABA 1h | 31.7419 | 10.9459 | 0.454022 | 0.156805 |
| ACC 1h | 39.1606 | 16.1838 | 0.448694 | 0.187175 |
| BABA 1h | 41.8721 | 20.4886 | 0.379692 | 0.421303 |
| Chitin 1h | 31.4008 | 12.8647 | 0.403824 | 0.206544 |
| Epi 1h | 33.6422 | 10.3591 | 0.487668 | 0.181747 |
| SA 1h | 39.9337 | 11.7517 | 0.491833 | 0.151997 |
| Me-JA 1h | 38.4248 | 12.1374 | 0.589433 | 0.220612 |
| Control 6h | 359.961 | 45.1071 | 4.98662 | 0.322955 |
| ABA 6h | 89.7186 | 11.3485 | 1.17326 | 0.103337 |
| ACC 6h | 156.654 | 43.2696 | 1.77518 | 0.560882 |
| BABA 6h | 273.673 | 17.4111 | 3.43771 | 0.255151 |
| Chitin 6h | 340.878 | 57.4999 | 4.40307 | 0.580124 |
| Epi 6h | 300.099 | 63.879 | 3.57171 | 0.562027 |
| SA 6h | 349.838 | 67.6235 | 4.7858 | 0.47892 |
| Me-JA 6h | 301.09 | 86.889 | 3.95664 | 0.8027 |
Source Transcript PGSC0003DMT400071278 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G39510.1 | +1 | 2e-46 | 164 | 102/304 (34%) | nodulin MtN21 /EamA-like transporter family protein | chr2:16491358-16493085 REVERSE LENGTH=374 |
