Probe CUST_31811_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31811_PI426222305 | JHI_St_60k_v1 | DMT400053120 | AGATCGATGAACTTTTTGCTTAGGAGGTGTTATAACCTTAGGTTATCTCCTATGTTTCAT |
All Microarray Probes Designed to Gene DMG400020607
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31811_PI426222305 | JHI_St_60k_v1 | DMT400053120 | AGATCGATGAACTTTTTGCTTAGGAGGTGTTATAACCTTAGGTTATCTCCTATGTTTCAT |
Microarray Signals from CUST_31811_PI426222305
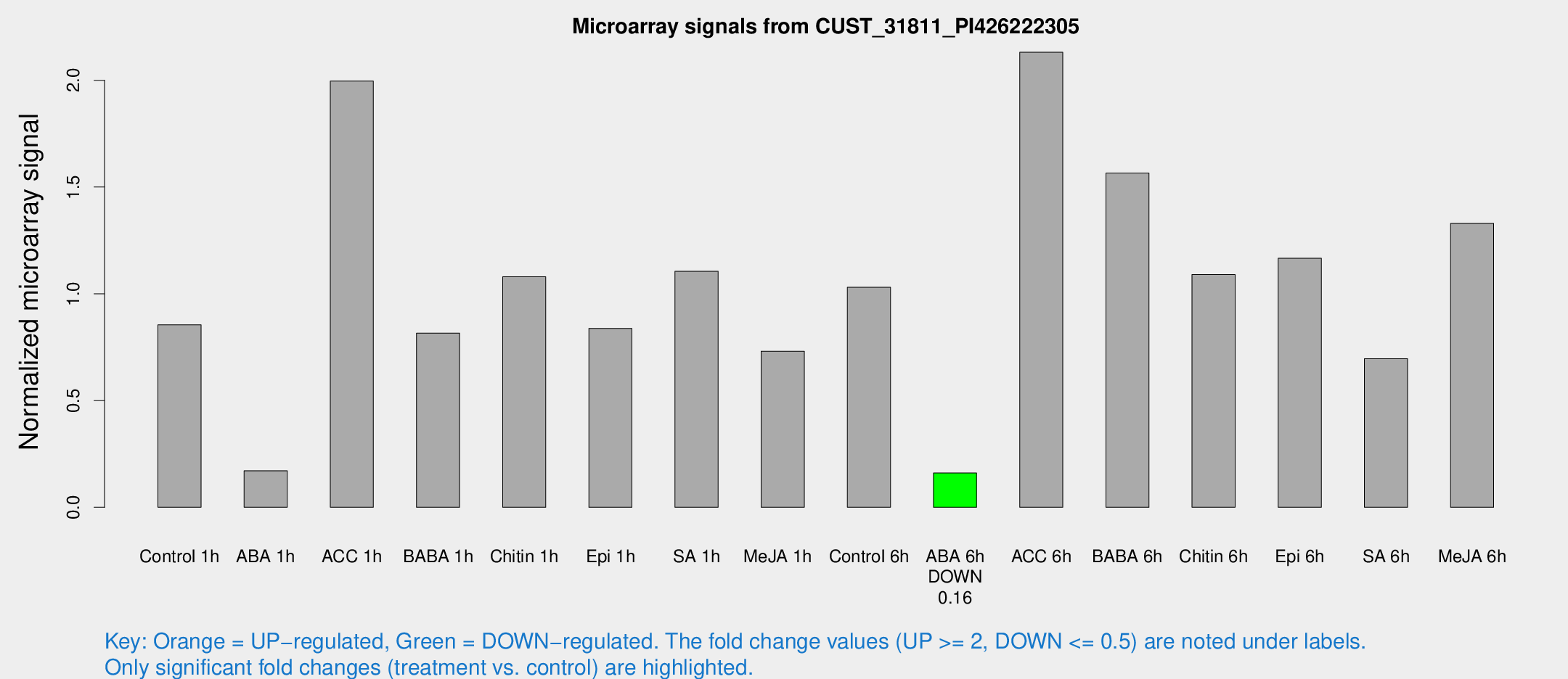
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.7835 | 16.402 | 0.854463 | 0.381499 |
| ABA 1h | 5.18483 | 3.00406 | 0.171333 | 0.099138 |
| ACC 1h | 76.6294 | 20.8825 | 1.99636 | 0.84313 |
| BABA 1h | 31.1711 | 10.2675 | 0.815891 | 0.271762 |
| Chitin 1h | 45.6163 | 25.4767 | 1.07999 | 0.835911 |
| Epi 1h | 31.7307 | 14.8722 | 0.837852 | 0.533567 |
| SA 1h | 46.5839 | 20.7644 | 1.10574 | 0.435508 |
| Me-JA 1h | 27.7435 | 11.1239 | 0.73119 | 0.54126 |
| Control 6h | 43.4994 | 15.9677 | 1.03026 | 0.478233 |
| ABA 6h | 5.84027 | 3.38429 | 0.160411 | 0.0928875 |
| ACC 6h | 93.553 | 32.9121 | 2.13154 | 0.408193 |
| BABA 6h | 66.4351 | 21.8463 | 1.56559 | 0.461244 |
| Chitin 6h | 46.4359 | 18.3151 | 1.09039 | 0.437646 |
| Epi 6h | 48.58 | 14.057 | 1.16621 | 0.351631 |
| SA 6h | 33.3902 | 18.3936 | 0.696157 | 0.43671 |
| Me-JA 6h | 56.979 | 28.3518 | 1.3299 | 0.726972 |
Source Transcript PGSC0003DMT400053120 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38310.1 | +1 | 7e-82 | 249 | 123/154 (80%) | PYR1-like 4 | chr2:16050251-16050874 FORWARD LENGTH=207 |
