Probe CUST_31718_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31718_PI426222305 | JHI_St_60k_v1 | DMT400082388 | GAACTTACTAATAAACTTCATACTGAAGGTGTTGTCATTCATTGGCATGGGATTCGACAG |
All Microarray Probes Designed to Gene DMG400032510
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31718_PI426222305 | JHI_St_60k_v1 | DMT400082388 | GAACTTACTAATAAACTTCATACTGAAGGTGTTGTCATTCATTGGCATGGGATTCGACAG |
Microarray Signals from CUST_31718_PI426222305
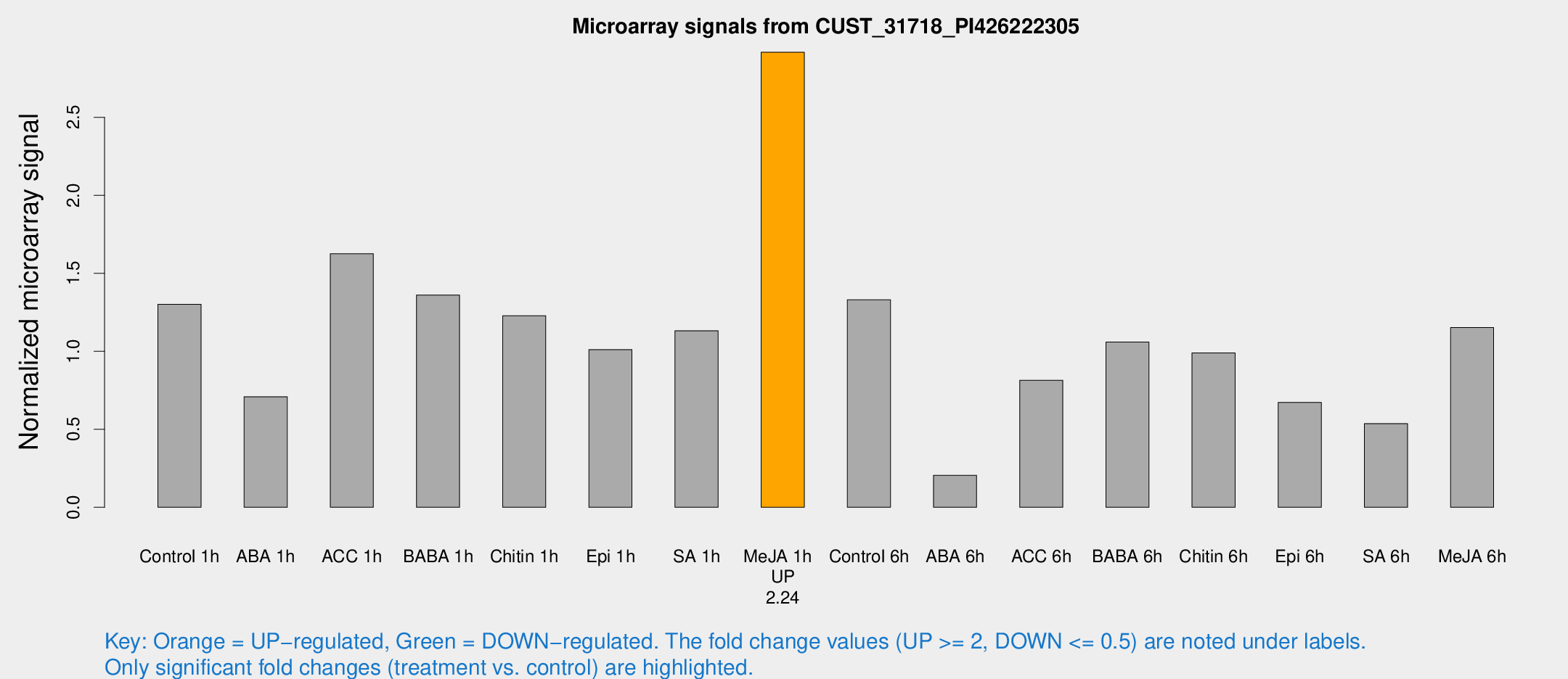
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 97.5735 | 26.007 | 1.30105 | 0.252018 |
| ABA 1h | 44.351 | 4.24462 | 0.708885 | 0.0683087 |
| ACC 1h | 121.674 | 21.9825 | 1.62498 | 0.202246 |
| BABA 1h | 93.7814 | 12.7561 | 1.36053 | 0.0964151 |
| Chitin 1h | 85.3576 | 27.9608 | 1.22819 | 0.410515 |
| Epi 1h | 61.9206 | 5.69082 | 1.01117 | 0.0810146 |
| SA 1h | 88.2182 | 25.9598 | 1.13189 | 0.330204 |
| Me-JA 1h | 169.517 | 19.9861 | 2.91709 | 0.297391 |
| Control 6h | 117.964 | 43.5523 | 1.32986 | 0.700128 |
| ABA 6h | 20.1913 | 10.8701 | 0.205333 | 0.144724 |
| ACC 6h | 69.4996 | 17.561 | 0.81458 | 0.0884268 |
| BABA 6h | 83.8904 | 7.46468 | 1.05899 | 0.0811554 |
| Chitin 6h | 75.5732 | 11.3551 | 0.989858 | 0.114945 |
| Epi 6h | 64.1304 | 26.0526 | 0.672742 | 0.391753 |
| SA 6h | 41.2507 | 13.8833 | 0.536534 | 0.160325 |
| Me-JA 6h | 94.0367 | 36.2506 | 1.15245 | 0.454146 |
Source Transcript PGSC0003DMT400082388 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G21105.1 | +3 | 0.0 | 768 | 368/549 (67%) | Plant L-ascorbate oxidase | chr5:7172727-7177409 FORWARD LENGTH=588 |
