Probe CUST_31582_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31582_PI426222305 | JHI_St_60k_v1 | DMT400073317 | CTTCAGTCAGCTAGCGTAGAGGCAAATCAAACAATCTTATATTAGTGGTTTAAGTATGAT |
All Microarray Probes Designed to Gene DMG400028491
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31582_PI426222305 | JHI_St_60k_v1 | DMT400073317 | CTTCAGTCAGCTAGCGTAGAGGCAAATCAAACAATCTTATATTAGTGGTTTAAGTATGAT |
Microarray Signals from CUST_31582_PI426222305
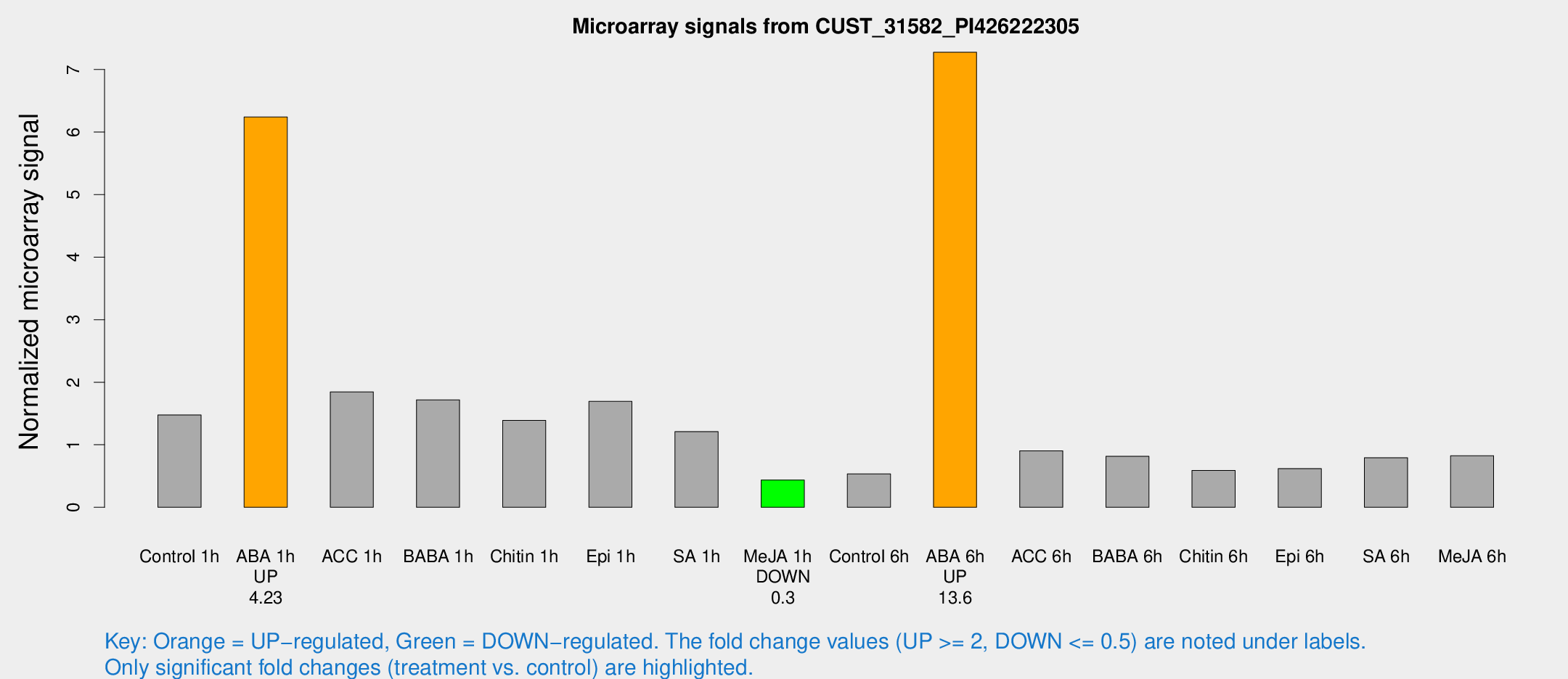
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 486.067 | 28.3456 | 1.47609 | 0.0887524 |
| ABA 1h | 1947.28 | 461.483 | 6.24109 | 1.50557 |
| ACC 1h | 680.857 | 184.039 | 1.84594 | 0.463956 |
| BABA 1h | 586.841 | 143.598 | 1.71889 | 0.324109 |
| Chitin 1h | 466.501 | 168.014 | 1.39 | 0.464971 |
| Epi 1h | 490.575 | 68.0951 | 1.69356 | 0.237899 |
| SA 1h | 466.838 | 179.396 | 1.21046 | 0.390487 |
| Me-JA 1h | 120.424 | 21.0512 | 0.43643 | 0.0414739 |
| Control 6h | 179.215 | 23.9778 | 0.535037 | 0.0596201 |
| ABA 6h | 2626.46 | 454.642 | 7.27643 | 0.969041 |
| ACC 6h | 341.59 | 20.2872 | 0.903654 | 0.0934587 |
| BABA 6h | 307.246 | 49.8039 | 0.81483 | 0.0994067 |
| Chitin 6h | 206.047 | 12.5454 | 0.588883 | 0.035842 |
| Epi 6h | 241.586 | 57.1544 | 0.6197 | 0.101748 |
| SA 6h | 261.814 | 32.7346 | 0.792324 | 0.047361 |
| Me-JA 6h | 289.572 | 76.4985 | 0.824495 | 0.154553 |
Source Transcript PGSC0003DMT400073317 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G25110.1 | +3 | 0.0 | 568 | 302/462 (65%) | CBL-interacting protein kinase 25 | chr5:8657740-8659206 REVERSE LENGTH=488 |
