Probe CUST_31532_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31532_PI426222305 | JHI_St_60k_v1 | DMT400042533 | GTGACTGACCAAATTTATATCCATCACCTCAGGATGTTTAATGTTGGTCTATGATGTATT |
All Microarray Probes Designed to Gene DMG400016499
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31532_PI426222305 | JHI_St_60k_v1 | DMT400042533 | GTGACTGACCAAATTTATATCCATCACCTCAGGATGTTTAATGTTGGTCTATGATGTATT |
Microarray Signals from CUST_31532_PI426222305
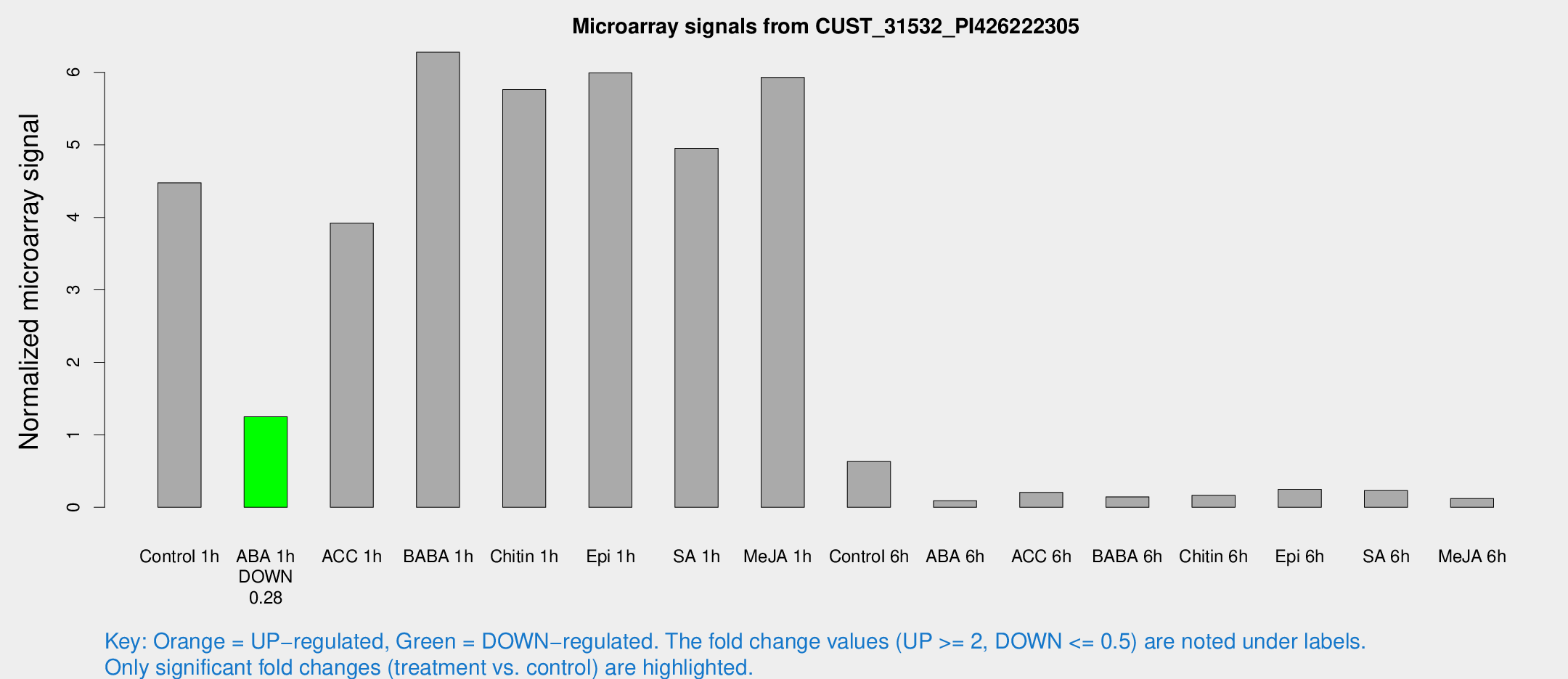
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 307.887 | 18.0174 | 4.47791 | 0.261833 |
| ABA 1h | 76.0016 | 5.18052 | 1.25017 | 0.0852195 |
| ACC 1h | 283.59 | 44.5217 | 3.92076 | 0.419692 |
| BABA 1h | 447.765 | 108.245 | 6.27805 | 1.1765 |
| Chitin 1h | 360.931 | 43.1418 | 5.76158 | 0.539811 |
| Epi 1h | 369.016 | 71.8827 | 5.99278 | 1.17313 |
| SA 1h | 392.036 | 134.484 | 4.95321 | 1.43824 |
| Me-JA 1h | 336.873 | 36.6661 | 5.9305 | 0.465235 |
| Control 6h | 45.3148 | 9.46053 | 0.631735 | 0.104288 |
| ABA 6h | 7.08735 | 3.11579 | 0.0912923 | 0.0447911 |
| ACC 6h | 22.1816 | 10.5934 | 0.20513 | 0.125276 |
| BABA 6h | 12.046 | 3.42698 | 0.144331 | 0.0521258 |
| Chitin 6h | 13.4342 | 4.40552 | 0.166552 | 0.051254 |
| Epi 6h | 23.8661 | 8.58112 | 0.248862 | 0.1887 |
| SA 6h | 16.8016 | 4.00468 | 0.229784 | 0.0565216 |
| Me-JA 6h | 8.49331 | 3.01353 | 0.121258 | 0.0448917 |
Source Transcript PGSC0003DMT400042533 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79770.1 | +3 | 5e-35 | 127 | 74/165 (45%) | Protein of unknown function (DUF1677) | chr1:30014371-30014874 FORWARD LENGTH=167 |
