Probe CUST_31324_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31324_PI426222305 | JHI_St_60k_v1 | DMT400071476 | AGGCTACTTGGCCATTGTTTACGTTCTACGCAAGTTTAACTTGTCGACGTGCTTCCTGAA |
All Microarray Probes Designed to Gene DMG401027811
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31324_PI426222305 | JHI_St_60k_v1 | DMT400071476 | AGGCTACTTGGCCATTGTTTACGTTCTACGCAAGTTTAACTTGTCGACGTGCTTCCTGAA |
| CUST_31392_PI426222305 | JHI_St_60k_v1 | DMT400071478 | CAGTTGGAGAACAGTACTCATTTCCAAAGTTATCCACAATAAAATCTTTGACAACTTTGG |
| CUST_31365_PI426222305 | JHI_St_60k_v1 | DMT400071477 | CTCACTAAAAACCGGAATCCCTGATTGGATATGGAAAATACCTCAACTAGAGTATCTGGA |
Microarray Signals from CUST_31324_PI426222305
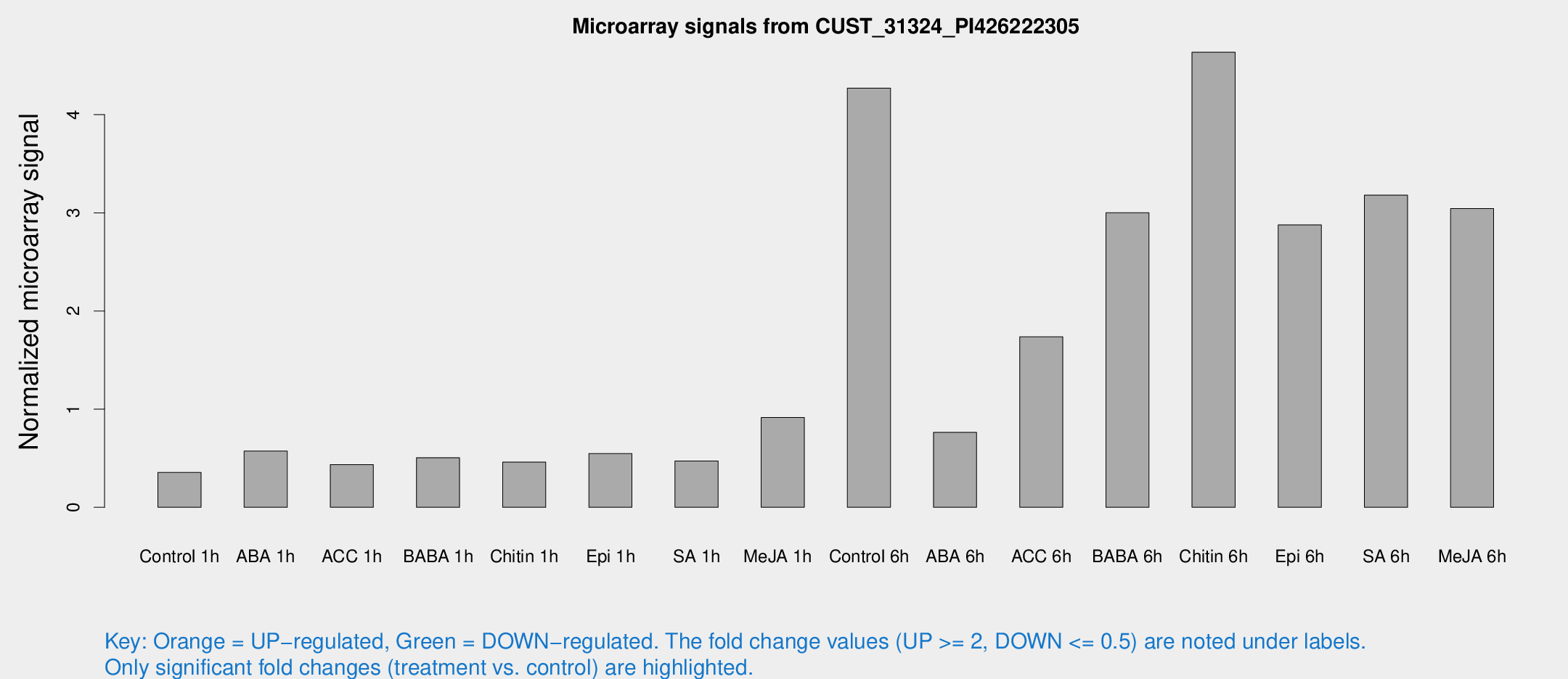
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.80073 | 3.37196 | 0.354447 | 0.205668 |
| ABA 1h | 9.41185 | 3.5442 | 0.572845 | 0.265797 |
| ACC 1h | 7.43919 | 4.26357 | 0.434076 | 0.241428 |
| BABA 1h | 8.18549 | 3.5703 | 0.505736 | 0.234907 |
| Chitin 1h | 6.85221 | 3.37014 | 0.46037 | 0.234506 |
| Epi 1h | 8.12629 | 3.3233 | 0.547372 | 0.251435 |
| SA 1h | 8.64974 | 3.43253 | 0.471285 | 0.224757 |
| Me-JA 1h | 12.327 | 3.54772 | 0.914275 | 0.267972 |
| Control 6h | 74.0191 | 16.5207 | 4.26966 | 0.719532 |
| ABA 6h | 14.4895 | 3.74417 | 0.763485 | 0.236888 |
| ACC 6h | 36.0687 | 10.1109 | 1.73698 | 0.46099 |
| BABA 6h | 58.5446 | 13.3618 | 3.00072 | 0.72271 |
| Chitin 6h | 121.57 | 71.7662 | 4.63638 | 4.66379 |
| Epi 6h | 55.8521 | 12.6513 | 2.87726 | 0.816051 |
| SA 6h | 65.1791 | 30.9489 | 3.18028 | 2.59063 |
| Me-JA 6h | 60.1758 | 25.7895 | 3.04357 | 1.2238 |
Source Transcript PGSC0003DMT400071476 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
