Probe CUST_31242_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31242_PI426222305 | JHI_St_60k_v1 | DMT400034927 | ATGGTGATGGTTTGCTCAATTTTGAGGAGTTTGTCATTATGATGCGTTGCTAGTTACATA |
All Microarray Probes Designed to Gene DMG400013427
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31242_PI426222305 | JHI_St_60k_v1 | DMT400034927 | ATGGTGATGGTTTGCTCAATTTTGAGGAGTTTGTCATTATGATGCGTTGCTAGTTACATA |
Microarray Signals from CUST_31242_PI426222305
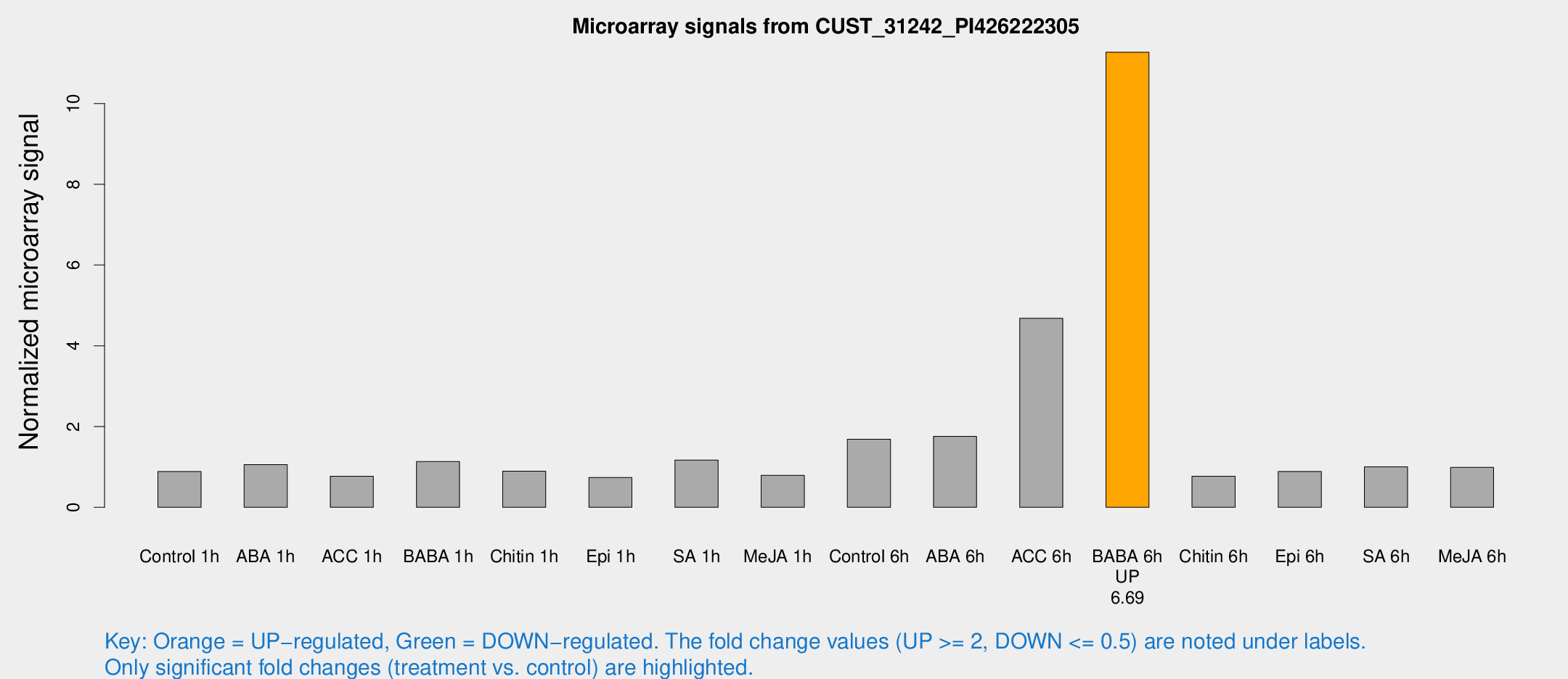
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.5435 | 6.16885 | 0.884258 | 0.451502 |
| ABA 1h | 13.6279 | 8.5014 | 1.05863 | 0.981726 |
| ACC 1h | 8.07533 | 3.46981 | 0.767245 | 0.349657 |
| BABA 1h | 11.7695 | 3.41062 | 1.13105 | 0.415728 |
| Chitin 1h | 8.70539 | 3.18193 | 0.894966 | 0.397105 |
| Epi 1h | 6.5744 | 3.1346 | 0.73932 | 0.371604 |
| SA 1h | 13.8857 | 4.38254 | 1.16847 | 0.476654 |
| Me-JA 1h | 6.81771 | 3.21579 | 0.792615 | 0.402203 |
| Control 6h | 21.4523 | 8.26403 | 1.68533 | 0.908159 |
| ABA 6h | 19.0203 | 3.53009 | 1.75519 | 0.345921 |
| ACC 6h | 56.7797 | 14.0161 | 4.68143 | 1.81543 |
| BABA 6h | 136.096 | 33.1255 | 11.2692 | 3.68098 |
| Chitin 6h | 8.199 | 3.58805 | 0.767995 | 0.339753 |
| Epi 6h | 10.2852 | 3.75487 | 0.88711 | 0.357682 |
| SA 6h | 10.5009 | 3.42374 | 1.00087 | 0.381826 |
| Me-JA 6h | 10.3684 | 3.23848 | 0.988591 | 0.346607 |
Source Transcript PGSC0003DMT400034927 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G76640.1 | +2 | 7e-24 | 95 | 69/135 (51%) | Calcium-binding EF-hand family protein | chr1:28765324-28765803 REVERSE LENGTH=159 |
